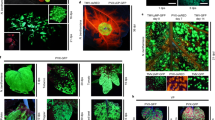
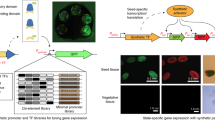
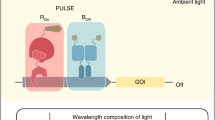
Abstract
Plant biotechnology predominantly relies on a restricted set of genetic parts with limited capability to customize spatiotemporal and conditional expression patterns. Synthetic gene circuits have the potential to integrate multiple customizable input signals through a processing unit constructed from biological parts to produce a predictable and programmable output. Here we present a suite of functional recombinase-based gene circuits for use in plants. We first established a range of key gene circuit components compatible with plant cell functionality. We then used these to develop a range of operational logic gates using the identify function (activation) and negation function (repression) in Arabidopsis protoplasts and in vivo, demonstrating their utility for programmable manipulation of transcriptional activity in a complex multicellular organism. Specifically, using recombinases and plant control elements, we activated transgenes in YES, OR and AND gates and repressed them in NOT, NOR and NAND gates; we also implemented the A NIMPLY B gate that combines activation and repression. Through use of genetic recombination, these circuits create stable long-term changes in expression and recording of past stimuli. This highly compact programmable gene circuit platform provides new capabilities for engineering sophisticated transcriptional programs and previously unrealized traits into plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All raw values of data presented in this study, output of statistical tests and summaries (including central tendency and variation), list of primer sequences and the sequences of the plasmids used in this study are available in a Zenodo repository64 with the identifier 10.5281/zenodo.6381286.
Code availability
R code used to perform statistical tests and generate plots is available in a Zenodo repository64 with the identifier 10.5281/zenodo.6381286.
References
Thompson, A. J. et al. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23, 363–374 (2000).
Iuchi, S. et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333 (2001).
Feeney, M., Frigerio, L., Cui, Y. & Menassa, R. Following vegetative to embryonic cellular changes in leaves of Arabidopsis overexpressing LEAFY COTYLEDON2. Plant Physiol. 162, 1881–1896 (2013).
Vanhercke, T. et al. Step changes in leaf oil accumulation via iterative metabolic engineering. Metab. Eng. 39, 237–246 (2017).
He, R. et al. Overexpression of 9-cis-epoxycarotenoid dioxygenase cisgene in grapevine increases drought tolerance and results in pleiotropic effects. Front. Plant Sci. 9, 970 (2018).
Brophy, J. A. N. Toward synthetic plant development. Plant Physiol. 188, 738–748 (2021).
Brophy, J. A. N., Magallon, K. J., Kniazev, K. & Dinneny, J. R. Synthetic genetic circuits enable reprogramming of plant roots. Preprint at https://www.biorxiv.org/content/10.1101/2022.02.02.478917v1 (2022).
Pires, N. D. et al. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl Acad. Sci. USA 110, 9571–9576 (2013).
Madrid, E., Chandler, J. W. & Coupland, G. Gene regulatory networks controlled by FLOWERING LOCUS C that confer variation in seasonal flowering and life history. J. Exp. Bot. 72, 4–14 (2021).
Setty, Y., Mayo, A. E., Surette, M. G. & Alon, U. Detailed map of a cis-regulatory input function. Proc. Natl Acad. Sci. USA 100, 7702–7707 (2003).
Krakauer, D. C., Müller, L., Prohaska, S. J. & Stadler, P. F. Design specifications for cellular regulation. Theory Biosci. 135, 231–240 (2016).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Lohmueller, J. J., Armel, T. Z. & Silver, P. A. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 40, 5180–5187 (2012).
Nevozhay, D., Zal, T. & Balázsi, G. Transferring a synthetic gene circuit from yeast to mammalian cells. Nat. Commun. 4, 1451 (2013).
Siuti, P., Yazbek, J. & Lu, T. K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 31, 448–452 (2013).
Gaber, R. et al. Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nat. Chem. Biol. 10, 203–208 (2014).
Roquet, N., Soleimany, A. P., Ferris, A. C., Aaronson, S. & Lu, T. K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).
Weinberg, B. H. et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 35, 453–462 (2017).
Müller, M. et al. Designed cell consortia as fragrance-programmable analog-to-digital converters. Nat. Chem. Biol. 13, 309–316 (2017).
Guiziou, S., Mayonove, P. & Bonnet, J. Hierarchical composition of reliable recombinase logic devices. Nat. Commun. 10, 456 (2019).
Zúñiga, A. et al. Rational programming of history-dependent logic in cellular populations. Nat. Commun. 11, 4758 (2020).
Bowyer, J. E., Ding, C., Weinberg, B. H., Wong, W. W. & Bates, D. G. A mechanistic model of the BLADE platform predicts performance characteristics of 256 different synthetic DNA recombination circuits. PLoS Comput. Biol. 16, e1007849 (2020).
Schreiber, T., Prange, A. & Tissier, A. F. Split-TALE—a TALE-based two-component system for synthetic biology applications in planta. Plant Physiol. 179, 1001–1012 (2019).
Bernabé-Orts, J. M. et al. A memory switch for plant synthetic biology based on the phage ϕC31 integration system. Nucleic Acids Res. 48, 3379–3394 (2020).
Lloyd, J. P. B. & Lister, R. Epigenome plasticity in plants. Nat. Rev. Genet. 23, 55–68 (2022).
Jones, J. M. & Gellert, M. The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 200, 233–248 (2004).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Engler, C. et al. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843 (2014).
Diamos, A. G. & Mason, H. S. Chimeric 3′ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J. 16, 1971–1982 (2018).
Andreou, A. I., Nirkko, J., Ochoa-Villarreal, M. & Nakayama, N. Mobius Assembly for Plant Systems highlights promoter–terminator interaction in gene regulation. Preprint at https://www.biorxiv.org/content/10.1101/2021.03.31.437819v1 (2021).
Efroni, I. et al. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165, 1721–1733 (2016).
Wu, F.-H. et al. Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Schaumberg, K. A. et al. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat. Methods 13, 94–100 (2016).
Padidam, M. & Cao, Y. Elimination of transcriptional interference between tandem genes in plant cells. Biotechniques 31, 328–330, 332–334 (2001).
Nagaya, S., Kawamura, K., Shinmyo, A. & Kato, K. The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332 (2010).
Rayson, S. et al. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS ONE 7, e31917 (2012).
Lloyd, J. P. B. & Davies, B. SMG1 is an ancient nonsense-mediated mRNA decay effector. Plant J. 76, 800–810 (2013).
Causier, B., Hopes, T., McKay, M., Paling, Z. & Davies, B. Plants utilise ancient conserved peptide upstream open reading frames in stress-responsive translational regulation. Plant Cell Environ. 45, 1229–1241 (2022).
Sanfaçon, H. & Hohn, T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature 346, 81–84 (1990).
Han, Y.-J., Kim, Y.-M., Hwang, O.-J. & Kim, J.-I. Characterization of a small constitutive promoter from Arabidopsis translationally controlled tumor protein (AtTCTP) gene for plant transformation. Plant Cell Rep. 34, 265–275 (2015).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6, e16765 (2011).
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S. & Somerville, C. R. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl Acad. Sci. USA 97, 3718–3723 (2000).
Heidstra, R., Welch, D. & Scheres, B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969 (2004).
Vergunst, A. C., Jansen, L. E. & Hooykaas, P. J. Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res. 26, 2729–2734 (1998).
Vergunst, A. C. & Hooykaas, P. J. Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol. Biol. 38, 393–406 (1998).
Sieburth, L. E., Drews, G. N. & Meyerowitz, E. M. Non-autonomy of AGAMOUS function in flower development: use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development 125, 4303–4312 (1998).
Marquès-Bueno, M. D. M. et al. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 85, 320–333 (2016).
Craft, J. et al. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41, 899–918 (2005).
Weinberg, B. H. et al. High-performance chemical- and light-inducible recombinases in mammalian cells and mice. Nat. Commun. 10, 4845 (2019).
Odell, J., Caimi, P., Sauer, B. & Russell, S. Site-directed recombination in the genome of transgenic tobacco. Mol. Gen. Genet. 223, 369–378 (1990).
Russell, S. H., Hoopes, J. L. & Odell, J. T. Directed excision of a transgene from the plant genome. Mol. Gen. Genet. 234, 49–59 (1992).
Schürholz, A.-K. et al. A comprehensive toolkit for inducible, cell type-specific gene expression in Arabidopsis. Plant Physiol. 178, 40–53 (2018).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Logemann, E., Birkenbihl, R. P., Ülker, B. & Somssich, I. E. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2, 16 (2006).
Shimada, T. L., Shimada, T. & Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010).
Engler, C., Gruetzner, R., Kandzia, R. & Marillonnet, S. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553 (2009).
Patron, N. J. et al. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol. 208, 13–19 (2015).
Libiakova, G., Jørgensen, B., Palmgren, G., Ulvskov, P. & Johansen, E. Efficacy of an intron-containing kanamycin resistance gene as a selectable marker in plant transformation. Plant Cell Rep. 20, 610–615 (2001).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Naseer, S. et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl Acad. Sci. USA 109, 10101–10106 (2012).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Lloyd, J. P. B. et al. Synthetic memory circuits for programmable cell reconfiguration in plants. https://doi.org/10.5281/zenodo.6381286 (2022).
Acknowledgements
We would like to thank B. Johnston for advice on R code; M. Oliva and D. Collings for help with confocal image analysis; I. Roux for suggestion of the in vitro recombinase assay; Y.-H. Chooi for the kind donation of purified recombinant Cre enzyme; B. Crawford for the kind donation of a pOp6 promoter-containing plasmid; C. Helliwell for graciously providing the LhGR seeds used in this study; and W. Wong for generous donation of mammalian BLADE plasmids. The plasmid sequencing data were generated on instrumentation supported by the Australian Cancer Research Foundation Centre for Advanced Cancer Genomics and Genomics WA. This work was supported by the following grants to R.L.: Australian Research Council (ARC) Centre of Excellence in Plant Energy Biology (CE140100008), ARC DP210103954, NHMRC Investigator Grant GNT1178460, Silvia and Charles Viertel Senior Medical Research Fellowship and Howard Hughes Medical Institute International Research Scholarship. T.S. was supported by the Hackett Postgraduate Research Scholarship. M.A.K. was supported by an International Postgraduate Research Scholarship. B.K. was supported by the CSIRO Synthetic Biology Future Science Platform.
Author information
Authors and Affiliations
Contributions
J.P.B.L. and R.L. conceived of the project and wrote the manuscript. J.P.B.L., with F.L. and P.G., conducted the experiments, with assistance from B.K. and E.F. J.P., T.S. and C.P. conducted the plasmid sequencing. M.A.K. developed the protoplast transformation protocol. M.A.K. and B.K. provided technical assistance with the protoplast assay, luciferase assay and cloning. All authors approved of and contributed to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Diego Orzaez, Anna Stepanova and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and uncropped gels for Supplementary Fig. 2
Rights and permissions
About this article
Cite this article
Lloyd, J.P.B., Ly, F., Gong, P. et al. Synthetic memory circuits for stable cell reprogramming in plants. Nat Biotechnol 40, 1862–1872 (2022). https://doi.org/10.1038/s41587-022-01383-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01383-2
This article is cited by
-
Choreographing root architecture and rhizosphere interactions through synthetic biology
Nature Communications (2024)
-
An orthogonalized PYR1-based CID module with reprogrammable ligand-binding specificity
Nature Chemical Biology (2024)
-
Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts
Nature Communications (2024)
-
Development of new binary expression systems for plant synthetic biology
Plant Cell Reports (2024)
-
Science fosters ongoing reassessments of plant capabilities
Theoretical and Experimental Plant Physiology (2024)