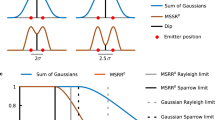
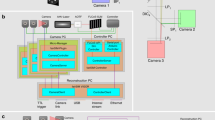
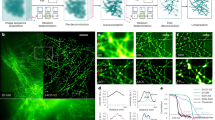
Abstract
A main determinant of the spatial resolution of live-cell super-resolution (SR) microscopes is the maximum photon flux that can be collected. To further increase the effective resolution for a given photon flux, we take advantage of a priori knowledge about the sparsity and continuity of biological structures to develop a deconvolution algorithm that increases the resolution of SR microscopes nearly twofold. Our method, sparse structured illumination microscopy (Sparse-SIM), achieves ~60-nm resolution at a frame rate of up to 564 Hz, allowing it to resolve intricate structures, including small vesicular fusion pores, ring-shaped nuclear pores formed by nucleoporins and relative movements of inner and outer mitochondrial membranes in live cells. Sparse deconvolution can also be used to increase the three-dimensional resolution of spinning-disc confocal-based SIM, even at low signal-to-noise ratios, which allows four-color, three-dimensional live-cell SR imaging at ~90-nm resolution. Overall, sparse deconvolution will be useful to increase the spatiotemporal resolution of live-cell fluorescence microscopy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw images from Figs. 1, 4a,m,j and 6e, and Extended Data Fig. 6, and several templates for processing different types of structures along with example datasets can be found at https://github.com/WeisongZhao/Sparse-SIM/releases/tag/v1.0.3 or https://doi.org/10.5281/zenodo.5079743. All other data that support the findings of this study are available from the corresponding author on request.
Code availability
Our light-weight MATLAB framework for video production is available at https://github.com/WeisongZhao/img2vid. Our adaptive filter has been written as an ImageJ plug-in and can be found at https://github.com/WeisongZhao/AdaptiveMedian.imagej. The version of sparse deconvolution software used in this manuscript (accompanied with a user manual) is available as Supplementary Software. The updating version of readily usable executable binary files for Windows (as ‘.exe’) and Mac (as ‘.app’) operating systems can be found at https://github.com/WeisongZhao/Sparse-SIM/releases/tag/v1.0.3, and the corresponding source code can be found at https://github.com/WeisongZhao/Sparse-SIM.
References
Nixon-Abell, J. et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, aaf3928 (2016).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017).
Hess, S. T. et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl Acad. Sci. USA 104, 17370–17375 (2007).
Shroff, H., Galbraith, C. G., Galbraith, J. A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Westphal, V. et al. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science 320, 246–249 (2008).
Zhu, L., Zhang, W., Elnatan, D. & Huang, B. Faster STORM using compressed sensing. Nat. Methods 9, 721–723 (2012).
Shin, W. et al. Visualization of membrane pore in live cells reveals a dynamic-pore theory governing fusion and endocytosis. Cell 173, 934–945 (2018).
Godin, A. G., Lounis, B. & Cognet, L. Super-resolution microscopy approaches for live cell imaging. Biophys. J. 107, 1777–1784 (2014).
Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 36, 451–459 (2018).
Li, D. et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 (2015).
Guo, Y. et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175, 1430–1442 (2018).
Wolter, H. On Basic Analogies and Principal Differences Between Optical and Electronic Information, Vol. 1 (Elsevier, 1961).
Harris, J. L. Diffraction and resolving power. J. Opt. Soc. Am. 54, 931–936 (1964).
Goodman, J. W. Introduction to Fourier Optics (Roberts and Company Publishers, 2005).
Lindberg, J. Mathematical concepts of optical superresolution. J. Opt. 14, 083001 (2012).
Bertero, M. & De Mol, C. Super-Resolution by Data Inversion, Vol. 36 (Elsevier, 1996).
Richardson, W. H. Bayesian-based iterative method of image restoration. J. Opt. Soc. Am. 62, 55–59 (1972).
Lucy, L. B. An iterative technique for the rectification of observed distributions. Astron. J. 79, 745 (1974).
Lucy, L. B. Resolution limits for deconvolved images. Astron. J. 104, 1260–1265 (1992).
Puschmann, K. G. & Kneer, F. On super-resolution in astronomical imaging. Astron. Astrophys. 436, 373–378 (2005).
Gazit, S., Szameit, A., Eldar, Y. C. & Segev, M. Super-resolution and reconstruction of sparse sub-wavelength images. Opt. Express 17, 23920–23946 (2009).
Demanet, L. & Nguyen, N. The recoverability limit for superresolution via sparsity. Preprint at https://arxiv.org/abs/1502.01385 (2015).
Fannjiang, A. C. Compressive imaging of subwavelength structures. SIAM J. Imaging Sci. 2, 1277–1291 (2009).
Schulz, O. et al. Resolution doubling in fluorescence microscopy with confocal spinning-disk image scanning microscopy. Proc. Natl Acad. Sci. USA 110, 21000–21005 (2013).
Zong, W. et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017).
Sun, D.-E. et al. Click-ExM enables expansion microscopy for all biomolecules. Nat. Methods 18, 107–113 (2021).
Dey, N. et al. Richardson–Lucy algorithm with total variation regularization for 3D confocal microscope deconvolution. Microsc. Res. Tech. 69, 260–266 (2006).
Laasmaa, M., Vendelin, M. & Peterson, P. Application of regularized Richardson–Lucy algorithm for deconvolution of confocal microscopy images. J. Microsc. 243, 124–140 (2011).
Candes, E. J. & Tao, T. Near-optimal signal recovery from random projections: universal encoding strategies? IEEE Trans. Inf. Theory 52, 5406–5425 (2006).
Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril of deep learning in microscopy. Nat. Methods 18, 131–132 (2021).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019).
Gu, L. et al. Molecular resolution imaging by repetitive optical selective exposure. Nat. Methods 16, 1114–1118 (2019).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013).
Ma, J., Kelich, J. M., Junod, S. L. & Yang, W. Super-resolution mapping of scaffold nucleoporins in the nuclear pore complex. J. Cell Sci. 130, 1299–1306 (2017).
Gottfert, F. et al. Strong signal increase in STED fluorescence microscopy by imaging regions of subdiffraction extent. Proc. Natl Acad. Sci. USA 114, 2125–2130 (2017).
Xia, S. et al. Nanoscale architecture of the cortical actin cytoskeleton in embryonic stem cells. Cell Rep. 28, 1251–1267 (2019).
Szameit, A. et al. Sparsity-based single-shot subwavelength coherent diffractive imaging. Nat. Mater. 11, 455–459 (2012).
Nieuwenhuizen, R. P. et al. Measuring image resolution in optical nanoscopy. Nat. Methods 10, 557–562 (2013).
Culley, S. et al. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat. Methods 15, 263–266 (2018).
Ornberg, R. L. & Reese, T. S. Beginning of exocytosis captured by rapid-freezing of Limulus amebocytes. J. Cell Biol. 90, 40–54 (1981).
York, A. G. et al. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat. Methods 10, 1122–1126 (2013).
York, A. G. et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat. Methods 9, 749–754 (2012).
Muller, C. B. & Enderlein, J. Image scanning microscopy. Phys. Rev. Lett. 104, 198101 (2010).
Theer, P., Mongis, C. & Knop, M. PSFj: know your fluorescence microscope. Nat. Methods 11, 981–982 (2014).
Saffarian, S., Cocucci, E. & Kirchhausen, T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 7, e1000191 (2009).
Luisier, F., Vonesch, C., Blu, T. & Unser, M. Fast interscale wavelet denoising of Poisson-corrupted images. Signal Process. 90, 415–427 (2010).
Chu, B. B. et al. Cholesterol transport through lysosome–peroxisome membrane contacts. Cell 161, 291–306 (2015).
Wang, Y., Yang, J., Yin, W. & Zhang, Y. A new alternating minimization algorithm for total variation image reconstruction. SIAM J. Imaging Sci. 1, 248–272 (2008).
Xue, J. Z. & Funabiki, H. Nuclear assembly shaped by microtubule dynamics. Nucleus 5, 40–46 (2014).
Cox, C. I. & Sheppard, C. Information capacity and resolution in an optical system. J. Opt. Soc. Am. A 3, 1152–1158 (1986).
Descloux, A., Grußmayer, K. S. & Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 16, 918–924 (2019).
Zhang, Y. et al. Mitochondria determine the sequential propagation of the calcium macrodomains revealed by the super-resolution calcium lantern imaging. Sci. China Life Sci. 63, 1543–1551 (2020).
Aaron, J. & Chew, T. -L. A guide to accurate reporting in digital image processing—can anyone reproduce your quantitative analysis? J. Cell Sci. 134, jcs254151 (2021).
Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Kim, D.-N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Tillberg, P. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Zhang, Z., Nishimura, Y. & Kanchanawong, P. Extracting microtubule networks from superresolution single-molecule localization microscopy data. Mol. Biol. Cell 28, 333–345 (2017).
De Vries, F. P. Automatic, adaptive, brightness independent contrast enhancement. Signal Process. 21, 169–182 (1990).
Meyer, F. & Beucher, S. Morphological segmentation. J. Vis. Commun. Image Represent. 1, 21–46 (1990).
Yanowitz, S. D. & Bruckstein, A. M. A new method for image segmentation. Comput. Gr. Image Process. 46, 82–95 (1989).
Peng, T. et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8, 14836 (2017).
Geissbuehler, M. & Lasser, T. How to display data by color schemes compatible with red–green color perception deficiencies. Opt. Express 21, 9862–9874 (2013).
Royer, L. A. et al. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods 12, 480–481 (2015).
Schmid, B. et al. 3Dscript: animating 3D/4D microscopy data using a natural-language-based syntax. Nat. Methods 16, 278–280 (2019).
Acknowledgements
We thank B. Hille and C. Xu for their reading and critical comments on the manuscript. We thank M. Knop and P. Theer for their feedback and valuable discussions of the bead correction factor and C. Zhang and J. Ma for the sharing of nuclear pore vectors. We thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with STED imaging experiments. L.C. acknowledges support by grants from the National Natural Science Foundation of China (nos 92054301, 81925022, 31821091 and 91750203), the National Science and Technology Major Project Program (no. 2016YFA0500400) and the Beijing Natural Science Foundation (no. Z20J00059). H.L. acknowledges support by grants from the National Natural Science Foundation of China (no. 61805057), the Young Elite Scientists Sponsorship Program (no. 2018QNRC001) and the Natural Science Foundation of Heilongjiang Province (YQ2021F013). L.C. acknowledges support by the High-Performance Computing Platform of Peking University. H.L. and J.L. acknowledge support by the State Key Laboratory of Robotics and Systems. S. Zhao acknowledges support by the Boya Postdoctoral Fellowship of Peking University. H.M. acknowledges support by grants from the National Natural Science Foundation of China (no. 32071458). Y.L. acknowledges support by grants from the National Natural Science Foundation of China (no. 91854112).
Author information
Authors and Affiliations
Contributions
L.C. and H.L. supervised the project. W.Z., H.L., L.C. and X.H. initiated and conceived the research. W.Z. developed the algorithm and implemented the corresponding software with the contribution and under the supervision of H.L. L.L., X.H. and S.X. performed the SIM experiments. S. Zhao and Y.Z. performed the SD-SIM experiments. S.Z. and C.S. performed the STED experiments. R.W. performed the MTPM experiments. L.G. performed the ROSE experiments under the supervision of W.J. Y.S. and S. Zhang prepared the DNA origami samples under the supervision of B.D. and W.J., respectively. D.S. prepared the expansion samples under the supervision of X.C. W.Z. performed the simulations and the theoretical analysis with contributions from G.Q., Z.H., J.W., R.C. and Y.M. W.Z. analyzed the data and prepared the figures and videos with contributions from S. Zhao and L.L. B.-L.S. and J.X. provided some of the reagents and participated in some of the discussions. Y.L., H.M., J.T., J.L. and B.-L.S. participated in discussions during the development of the manuscript. L.C., H.L. and W.Z. wrote the manuscript with input from all authors. All of the authors participated in discussions and data interpretation.
Corresponding authors
Ethics declarations
Competing interests
L.C., H.L., W.Z. and X.H. have a pending patent application on the presented framework.
Additional information
Peer review information Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Flowchart of the sparse deconvolution.
Raw images from 2D-SIM or SD-SIM were background subtracted (optional operation), upsampled (optional operation), and reconstructed with the sparsity and the continuity a priori information along the xy-t (z) axes before the final iterative deconvolution. Experiments were repeated ten times independently with similar results. Scale bars: 1 μm.
Extended Data Fig. 2 Sparse deconvolution for Argo-SIM slide under different SNR conditions.
(a-e) The results with different SNRs. We set the imaging condition of results in (a) as the full SNR (‘1 SNR’), and the SNRs decreased from 1/2 to 1/16 for the results in (b-e). Left panel, from top to bottom: the averaged wide-field (WF) image (NA 1.4), the image reconstructed by Wiener-SIM (‘2D-SIM’), the image reconstructed by Wiener-SIM and followed by RL deconvolution with 50 iterations (‘RL-SIM’), and our Sparse-SIM model result. Right panel, from top to bottom: the corresponding fluorescence profiles along lines crossing the middle of the left panel. When the SNR was in the range of 1~1/4, the 2D-SIM, RL-SIM, and Sparse-SIM distinguished pair lines up to 150 nm, 120 nm, and 60 nm, respectively. When the SNR was in the range of 1/8~1/16, 2D-SIM and RL-SIM failed to reconstruct the paired parallel lines, and snowflake-like artifacts emerged. However, under similar conditions, Sparse-SIM still resolved the pair parallel lines up to 90 nm and 120 nm, respectively.
Extended Data Fig. 3 Analysis of possible artifacts in the caveolae images reconstructed with the sparse deconvolution, which was further compared with other deconvolution methods.
(a) A representative COS-7 cell labeled with caveolin-EGFP under TIRF-SIM and Sparse-SIM ×2 (whole FOV from Fig. 4j). (b-e) The region enclosed by the white box in (a) was magnified and shown under the non-iterative method (TIRF-SIM), sparse deconvolution (Sparse-SIM ×2), or sparse deconvolution followed by convolving back with the resolution scaled function (RSF). The RSF is estimated between TIRF-SIM and Sparse-SIM ×2, and the FWHM of this estimated RSF is 78 nm. (e) The resolution scaled error (RSE) map of Sparse-SIM ×2 against the raw TIRF-SIM image. Before the RSE map estimation, the intensity of TIRF-SIM and Sparse-SIM images are normalized to the range of 0~1, and the corresponding residual image (RSE map) is color-coded within the range of 0~1. (f) Magnified views in (b-e). (g) The region enclosed by the white rectangle in (f) was magnified and reconstructed with a non-iterative method (TIRF-SIM), followed by image squares (TIRF-SIM square), or Fourier interpolated followed by RL-deconvolution (TIRF-SIM + RL ×2) for 3 or 25 iterations, or Fourier interpolated followed by the sparse deconvolution (Sparse-SIM ×2). Experiments were repeated five times independently with similar results. Scale bars: (a-e) 2 μm; (f) 1 μm; (g) 100 nm.
Extended Data Fig. 4 Relative movements between sub-organellar structures observed by dual-color Sparse-SIM.
(a) The inner and outer mitochondrial membranes (OMMs and IMMs) labeled with Tom20–mCherry and Mito-Tracker Green in a live COS-7 cell. (b) Magnified views from the white box in (a). (c) Intensity profiles of OMMs (magenta) and IMMs (cyan) along the continuous and dashed lines in (b), with mitochondrial configurations shown in the right. Sparse-SIM readily detected two types of OMMs:IMMs configurations: a long crista extended from one side to ~142 nm away from the other side of the OMMs, and a short crista extended only ~44 nm towards the other side of the OMMs (Supplementary Video 7). (d) Average FRC resolutions (n = 10). (e) The white dashed box in (a) is enlarged and shown at three time points. It revealed rare events: the IMMs extension not being enclosed within the Tom20-labeled structures in a few frames. This result might be explained by the non-homogenous distribution of Tom20 protein on the OMMs. (f) A representative example of both the IMMs (cyan) and ER (magenta, Sec61β-mCherry). As shown in the inset, we found that ER tubules randomly contacted the mitochondria with equal probability at both the cristae regions and the regions between cristae. (g) Magnified views from the white box in (f). We found that the contact between one ER tubule and the top of a mitochondrion not only correlated with the directional movement of the latter but also the synergistically rearranged orientations of cristae thereafter (Supplementary Video 8). Centerline, medians; limits, 75% and 25%; whiskers, maximum and minimum; error bars, s.e.m; Experiments were repeated five times independently with similar results. Scale bars: (a, f) 1 μm; axial: 0.2 arbitrary units (a.u.); lateral: 100 nm; (b, e and g) 500 nm.
Extended Data Fig. 5 Three-dimensional image stack of fluorescent beads under SD-SIM and Sparse SD-SIM.
(a,b) A maximum intensity projection (MIP) view (left) and a horizontal section (right) of fluorescent beads (100 nm in diameter) recorded by SD-SIM (a) and after the sparse deconvolution (b), respectively. Insets in the left-lower corner show a magnified view of the same fluorescent bead under different reconstruction methods. (c) The corresponding Gaussian fitted profiles in (a, left-lower corner), which indicate that the lateral FWHM of SD-SIM (red) and Sparse SD-SIM (blue) are 185 nm (calibrated resolution ~165 nm) and 110 nm (calibrated resolution ~90 nm), respectively (Supplementary Note 9.2). (d) Magnified horizontal sections from the white boxes in (a–b) are shown in the left and right panels, while the SD-SIM image is processed with a median filter to avoid a non-converged fitted result. (e) We used Gaussian functions to fit the intensity profiles along the axial direction of the fluorescent bead in (d), yielding axial resolutions of 484 nm and 266 nm for SD-SIM and Sparse SD-SIM, respectively. (f) The gradually improved axial resolution (FWHM) of a 100 nm bead while increasing the weight of sparsity. (g) Measuring the FWHM with fluorescent beads with a diameter of 45 nm. The fitted FWHMs (cross-sections between white arrows displayed with white profiles in the right) of SD-SIM and Sparse SD-SIM are 175 nm and 92 nm, respectively. As shown with yellow profiles (cross-sections between yellow arrows), the Sparse SD-SIM resolved adjacent two beads with a distance of 95 nm. Experiments were repeated five times independently with similar results. Scale bars: (b) 4 μm, (b, inset) 100 nm and (d, f, g) 200 nm.
Extended Data Fig. 6 Dual-color live-cell imaging of clathrin and actin by Sparse SD-SIM (Supplementary Video 16).
(a,b) Color temporal projections of CCPs (a) and the actin filament network (b) recorded by SD-SIM (left) and Sparse SD-SIM (right) for 16.75 minutes at 5 s intervals. (c) CCPs (cyan) and the cortical actin cytoskeleton (magenta) in a COS-7 cell captured by Sparse SD-SIM. (d) Montages of the boxed region in (c) at five-time points are shown at a magnified scale; the first image observed under SD-SIM appears at the top left corner for comparison. It can be observed that a CCP docks stably at the junction of two actin filaments and then disappears from the focal plane as these neighboring filaments merge. Experiments were repeated five times independently with similar results. Scale bars: (a-c) 4 μm; (d) 500 nm.
Extended Data Fig. 7 ER-lysosome contacts revealed by the sparse SD-SIM.
(a) Contacts between ER tubules (labeled by Sec61-EGFP, green) and lysosomes (labeled by Lysotracker DeepRed, magenta) visualized by Sparse SD-SIM in live COS-7 cells (Supplementary Video 17). (b) Time-lapse images of typical lysosome-ER contact dynamics magnified from the dashed-boxed region in (a) by SD-SIM (top) and Sparse SD-SIM (bottom). The ER tubules moved along with the lysosome, followed by the contraction of the tubular structure to a polygon surrounding the lysosome (indicated by white arrows) and the final disappearance of the polygon due to the retraction of the ER tubules. Experiments were repeated five times independently with similar results. Scale bars: (a) 3 μm; (b) 2 μm.
Extended Data Fig. 8 Bona fide spatial resolution improvement of confocal and STED microscopes by the sparse deconvolution.
(a) Nuclear pores in HeLa cells were labeled with an anti-Mab414 primary antibody and the Alexa594 secondary antibody, and observed under the Confocal, Sparse-Confocal, STED, and Sparse-STED configurations. (b) Magnified views from the region enclosed in the white dashed box in (a) under different microscopes. Huygens- represents that the images were deconvoluted by Huygens Professional (Scientific Volume Imaging, The Netherlands). (c) The Fourier transforms of images obtained by the corresponding microscopes. Labeled with the corresponding decorrelation resolution. (d) A representative HeLa cell in which microtubules (green) and mitochondria (magenta) were labeled with anti-tubulin and anti-Tom20 primary antibodies. It was imaged under the Confocal, Sparse-Confocal, STED, and Sparse-STED configurations. (e) Magnified views from the region enclosed by the white dashed box in (d). (f) Resolutions are measured by the decorrelation method. S-Confocal: Sparse-Confocal; S-STED: Sparse-STED. Experiments were repeated five times independently with similar results. Scale bars: (a, c, d) 2 μm; (b) 200 nm; (e) 500 nm.
Extended Data Fig. 9 Extending the spatial resolution of a miniaturized two-photon microscope (MTPM) with sparse deconvolution.
(a) Three-dimensional distributions of neuronal dendrites and spines within a volume of 190 × 190 × 110 μm3 from the brain of a Thy1-GFP transgenic mouse were observed under the MTPM, and after RL (RL-MTPM) or the sparse deconvolution (Sparse-MTPM). Different focal planes away from the surface were color-coded and projected to one image (see Supplementary Video 18). (b-d) The xz views and their Fourier transforms (a) under different configurations (b, MTPM; c, RL-MTPM; d, Sparse-MTPM). (e) Magnified views from the region enclosed by the white box in (a) under different configurations (from left to right: MTPM, RL-MTPM, Sparse-MTPM). (f) Resolutions of designated configurations as calculated by the decorrelation method at different axial positions. Experiments were repeated five times independently with similar results. Scale bars: (a, d) 15 μm; (e) 3 μm.
Extended Data Fig. 10 Highly-correlated Ca2+ transients after the sparse deconvolution compared to the original data obtained by the SD-SIM.
(a, b) The representative COS-7 cell was transfected with GCaMP6s, stimulated with ATP (10 μM). One snapshot under the SD-SIM (a) and after the sparse deconvolution (b) were shown. (c) Magnified views of regions enclosed by white boxes 1-4 in (a, and b). (d) ATP stimulated calcium traces from corresponding macrodomains in (c). (e) Representative ATP stimulated whole-cell calcium traces. (f) ATP stimulated increases in fluorescence intensities of GCaMP6s from different macrodomains (4 cell × 4 regions) under the SD-SIM (x-axis) exhibited a linear relationship with those obtained under the Sparse SD-SIM microscope (y-axis). (g) Average minimal resolutions by the FRC method (n = 10). Centerline, medians; limits, 75% and 25%; whiskers, maximum and minimum; error bars, s.e.m. Experiments were repeated five times independently with similar results. Scale bars: (a) 5 μm; (c) 2 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1–41, Notes 1–10 and Tables 1–5.
Supplementary Software
This file contains the source codes of the Sparse deconvolution.
41587_2021_1092_MOESM4_ESM.mp4
Supplementary Video 1. Ring-shaped Nup98 pores resolved by Sparse-SIM 2×. Nuclear pores labeled with Nup98–GFP in a live COS-7 cell with 4-s intervals imaged by 2D-SIM, 2D-SIM followed by RL deconvolution and Sparse-SIM 2× (cf., Fig. 2c) demonstrate that only Sparse-SIM 2× can resolve the ring-shaped Nup98.
41587_2021_1092_MOESM5_ESM.mp4
Supplementary Video 2. Dense actin mesh network revealed due to improved spatial resolution and enhanced contrast. Part I compares the actin results by 2D-SIM, Hessian-SIM and Sparse-SIM, demonstrating that finer structures of the dense actin mesh network can be revealed only with Sparse-SIM (cf., Fig. 4a). Part II exhibits the corresponding profiles of the red and blue lines from 2D-SIM (left) and Sparse-SIM (right) over 20 frames at 5-s intervals, showing the enhanced contrast available with Sparse-SIM.
41587_2021_1092_MOESM6_ESM.mp4
Supplementary Video 3. Ring-shaped caveolae revealed by Sparse-SIM 2×. The caveolae in a COS-7 cell at 37 °C transfected with caveolin–EGFP visualized by TIRF, TIRF-SIM, Sparse-SIM and Sparse-SIM 2× (cf., Fig. 4j).
41587_2021_1092_MOESM7_ESM.mp4
Supplementary Video 4. Improved contrast of fluorescent vesicles deep in the cytosol under Sparse-SIM. In part I, the structures labeled with LAMP1–EGFP, LysoView and LipidSpot in COS-7 cells are shown from left to right at 5-s intervals (cf., Fig. 4m). Part II compares 2D-SIM, Hessian-SIM and Sparse-SIM performances on three individual organelles and the corresponding magnified regions of interest.
41587_2021_1092_MOESM8_ESM.mp4
Supplementary Video 5. Visualization of ultrafast vesicle fusion (played four times slower than real-world speed). Fusion pores by TIRF-SIM (top) and Sparse-SIM (bottom) played at a speed four times slower than real-world rates. Vesicles in an INS-1 cell labeled with VAMP2–pHluorin visualized by TIRF-SIM and Sparse-SIM, respectively (cf., Fig. 4o).
41587_2021_1092_MOESM9_ESM.mp4
Supplementary Video 6. Vesicle with a 60 nm pore diameter captured by Sparse-SIM at 564 Hz. The initial opening of the fusion pore occurred much earlier, and its duration was much longer under Sparse-SIM than under TIRF-SIM (same data as in Supplementary Video 5).
41587_2021_1092_MOESM10_ESM.mp4
Supplementary Video 7. Asynchronized movements between the OMMs and IMMs revealed by dual-color Sparse-SIM. OMMs and IMMs labeled by Tom20–mScarlet and MitoTracker Green, respectively, visualized by 2D-SIM and Sparse-SIM for 30 frames with a 0.2 s acquisition time (cf., Extended Data Fig. 4a). The extension of the IMM was not enclosed by the Tom20-labeled structures captured by Sparse-SIM in a few frames, demonstrating the non-homogenous distribution of Tom20 on the OMM.
41587_2021_1092_MOESM11_ESM.mp4
Supplementary Video 8. Relative dynamics between ER tubules and the IMM recorded by dual-color Sparse-SIM. The IMM (cyan) and ER (magenta) in a COS-7 cell labeled by MitoTracker Green and Sec61β–mCherry (cf., Extended Data Fig. 4f). Part I compares the IMM and ER by dual-color 2D-SIM and Sparse-SIM. In part II, the ER contacts the IMM, rearranging the orientations of the inner cristae structures. Part III shows the equal probability of ER tubules contacting the mitochondria at cristae regions or in the matrix between cristae.
41587_2021_1092_MOESM12_ESM.mp4
Supplementary Video 9. Ring-shaped structures of CCPs resolved by Sparse SD-SIM. Comparison of CCPs in COS-7 cells expressing clathrin–EGFP light chains for 100 time points with a 0.2 s exposure time imaged by SD-SIM and Sparse SD-SIM, respectively (cf., Fig. 5a). Disintegration (part II of the video) and disappearance (part III) events of CCPs are resolved only by Sparse SD-SIM.
41587_2021_1092_MOESM13_ESM.mp4
Supplementary Video 10. Four-color sub-90-nm resolution live-cell imaging of lysosomes, mitochondria, nuclei and microtubules. A COS-7 cell labeled with LAMP1–mCherry (yellow), MitoTracker (green), Hoechst (blue) and tubulin–EGFP (magenta) imaged by SD-SIM and Sparse SD-SIM for 100 frames at a 2.3 s exposure time (cf., Fig. 5h). Part I shows the SD-SIM performance of the four organelles, which gradually transfer to Sparse SD-SIM in sequence. Then, the four organelles under SD-SIM and Sparse SD-SIM are compared simultaneously showing the striking contrast. Part II displays the dynamics of the magnified selection region in part I.
41587_2021_1092_MOESM14_ESM.mp4
Supplementary Video 11. Live-cell 3D image of the OMM in a dividing cell ~7 µm in depth. Mitochondrial expression of the outer membrane marker Tom20–mCherry captured by SD-SIM and Sparse SD-SIM (cf., Fig. 5k). Part I shows the color-coded projection of the mitochondrial membrane in the z direction as the depth increases gradually and the y–z orthoslices along the x axis. Part II displays multiangle views of the 3D rendering volume of the mitochondrial membrane showing the details recorded by SD-SIM (red) and Sparse SD-SIM (green).
41587_2021_1092_MOESM15_ESM.mp4
Supplementary Video 12. ER tubular structures under a Nyquist-insufficient sampling paradigm resolved by Sparse SD-SIM 2×. Tubular ER in the periphery of a COS-7 cell labeled with Sec61β–EGFP. Part I shows a comparison of the tubular ER by SD-SIM, Hessian SD-SIM, Sparse SD-SIM and Sparse SD-SIM 2× (cf., Fig. 6a). Part II compares the tubular ER by SD-SIM (left) with those by Sparse SD-SIM (right) 2× for 500 time points at a 0.2 s acquisition time. A magnified view of the selected region in part II is shown in part III.
41587_2021_1092_MOESM16_ESM.mp4
Supplementary Video 13. Microtubules regulate lysosome–peroxisome membrane contacts in live cells captured by three-color Sparse SD-SIM 2× imaging. Part I shows a time-lapse full-field view comparison of SD-SIM and Sparse SD-SIM 2×. Part II shows the two zoomed-in examples showing a comparison of the images by SD-SIM and Sparse SD-SIM 2× (cf., Fig. 6e). The first example shows a lysosome moving along the microtubule to make membrane contact with the peroxisome for close to 2 min; the deformation of the lysosome membrane during the contact period can be detected only by Sparse SD-SIM 2×. The second example shows a lysosome–peroxisome membrane in contact moving along microtubules together in a relatively dense microtubule region; the movement can be detected only by Sparse SD-SIM 2×.
41587_2021_1092_MOESM17_ESM.mp4
Supplementary Video 14. Three-color 3D volume-rendered views under Sparse SD-SIM 2×. Live-cell three-color 3D imaging of a COS-7 cell labeled with tubulin–EGFP (green), Hoechst (cyan) and MitoTracker Deep Red FM (magenta) under Sparse SD-SIM 2× (cf., Fig. 6h). Part I shows x–y orthoslices gradually forming color-coded volumes of nuclei (left), mitochondria (middle) and microtubules (right). Part II displays three-color 3D volume-rendered views of nuclei, mitochondria and microtubules from various angles.
41587_2021_1092_MOESM18_ESM.mp4
Supplementary Video 15. Benchmarks of time-varying actin filaments under different approaches according to the ground truth 2D-SIM images. Two branched actin filaments were recorded by wide-field, wide-field + deconvolution, Sparse 2× and 2D-SIM (cf., Supplementary Fig. 31e). Using 2D-SIM as the ground truth, wide-field with Sparse 2× reveals fine structure, such as two branched actin filaments separated correctly.
41587_2021_1092_MOESM19_ESM.mp4
Supplementary Video 16. Relative dynamics of both actin filaments and CCPs captured by dual-color Sparse SD-SIM. Actin filaments (labeled by LifeAct–EGFP) and CCPs (labeled by clathrin–DsRed) in COS-7 cells for 200 time points at a 5-s interval (cf., Extended Data Fig. 6). A CCP stably docked at the intersection of two actin filaments that begins to disappear from the confocal plane as the neighboring filaments close up and meet (marked with a yellow circle).
41587_2021_1092_MOESM20_ESM.mp4
Supplementary Video 17. ER–lysosome contacts in live cells captured by dual-color Sparse SD-SIM. Part I shows the time-lapse full-field ER–lysosome (labeled with Sec61β–EGFP and Lysotracker Deep Red) view comparison of SD-SIM and Sparse SD-SIM (cf., Extended Data Fig. 7). Part II shows the zoom-in example showing typical lysosome–ER contact dynamics by SD-SIM and Sparse SD-SIM. Lysosome–ER contact dynamics can be clearly viewed under Sparse SD-SIM.
41587_2021_1092_MOESM21_ESM.mp4
Supplementary Video 18. Volumetric imaging of neuronal dendrites and spines by Sparse-MTPM. Three-dimensional distributions of neuronal dendrites and spines within a volume of 190 × 190 × 110 μm3 from the brain of a Thy1–GFP transgenic mouse observed with MTPM, RL-deconvoluted MTPM (RL-MTPM) and Sparse-MTPM (cf., Extended Data Fig. 9). Part I demonstrates x–y comparison of these three methods. Part II shows the x–y orthoslices gradually forming color-coded volumes of MTPM, RL-MTPM and Sparse-MTPM from left to right, respectively. Part III displays 3D volume-rendered views of neuronal dendrites and spines from various angles.
Rights and permissions
About this article
Cite this article
Zhao, W., Zhao, S., Li, L. et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat Biotechnol 40, 606–617 (2022). https://doi.org/10.1038/s41587-021-01092-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-01092-2
This article is cited by
-
Self-supervised denoising for multimodal structured illumination microscopy enables long-term super-resolution live-cell imaging
PhotoniX (2024)
-
StayGold variants for molecular fusion and membrane-targeting applications
Nature Methods (2024)
-
Noise learning of instruments for high-contrast, high-resolution and fast hyperspectral microscopy and nanoscopy
Nature Communications (2024)
-
Membrane transformations of fusion and budding
Nature Communications (2024)
-
Tracking endogenous proteins based on RNA editing-mediated genetic code expansion
Nature Chemical Biology (2024)