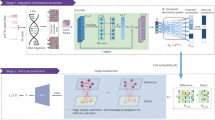
Abstract
Clinical applications of precision oncology require accurate tests that can distinguish true cancer-specific mutations from errors introduced at each step of next-generation sequencing (NGS). To date, no bulk sequencing study has addressed the effects of cross-site reproducibility, nor the biological, technical and computational factors that influence variant identification. Here we report a systematic interrogation of somatic mutations in paired tumor–normal cell lines to identify factors affecting detection reproducibility and accuracy at six different centers. Using whole-genome sequencing (WGS) and whole-exome sequencing (WES), we evaluated the reproducibility of different sample types with varying input amount and tumor purity, and multiple library construction protocols, followed by processing with nine bioinformatics pipelines. We found that read coverage and callers affected both WGS and WES reproducibility, but WES performance was influenced by insert fragment size, genomic copy content and the global imbalance score (GIV; G > T/C > A). Finally, taking into account library preparation protocol, tumor content, read coverage and bioinformatics processes concomitantly, we recommend actionable practices to improve the reproducibility and accuracy of NGS experiments for cancer mutation detection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All raw data (FASTQ files) are available on NCBI’s SRA database (SRP162370). The call set for somatic mutations in HCC1395, VCF files derived from individual WES and WGS runs, bam files for BWA-MEM alignments and source codes are available on NCBI’s ftp site (http://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/seqc/Somatic_Mutation_WG/).
Code availability
The code used to create figures and tables is deposited on GitHub under a BSD 2-Clause open-source license tagged at https://github.com/bioinform/somaticseq/tree/seqc2/utilities/Code_for_Figures/best_practices_manuscript. A snapshot can also be downloaded at https://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/seqc/Somatic_Mutation_WG/tools/.
References
Glasziou, P., Meats, E., Heneghan, C. & Shepperd, S. What is missing from descriptions of treatment in trials and reviews? Brit. Med. J. 336, 1472–1474 (2008).
Vasilevsky, N. A. et al. On the reproducibility of science: unique identification of research resources in the biomedical literature. PeerJ 1, e148 (2013).
Begley, C. G. & Ellis, L. M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Alioto, T. S. et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat. Commun. 6, 10001 (2015).
Griffith, M. et al. Genome Modeling System: a knowledge management platform for genomics. PLoS Comput. Biol. 11, e1004274 (2015).
Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017).
Xu, H., DiCarlo, J., Satya, R. V., Peng, Q. & Wang, Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics 15, 244 (2014).
Ghoneim, D. H., Myers, J. R., Tuttle, E. & Paciorkowski, A. R. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC Res. Notes 7, 864 (2014).
Wang, Q. et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 5, 91 (2013).
Simen, B. B. et al. Validation of a next-generation-sequencing cancer panel for use in the clinical laboratory. Arch. Pathol. Lab. Med. 139, 508–517 (2015).
Linderman, M. D. et al. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med. Genomics 7, 20 (2014).
Zook, J. M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 32, 246–251 (2014).
Zook, J. M. et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016).
Lin, M.-T. et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am. J. Clin. Pathol. 141, 856–866 (2014).
Singh, R. R. et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J. Mol. Diagn. 15, 607–622 (2013).
Griffith, M. et al. Optimizing cancer genome sequencing and analysis. Cell Syst. 1, 210–223 (2015).
Olson, N. D. et al. precisionFDA Truth Challenge V2: calling variants from short- and long-reads in difficult-to-map regions. Preprint at bioRxiv https://doi.org/10.1101/2020.11.13.380741 (2020).
Morrissy, A. S. et al. Spatial heterogeneity in medulloblastoma. Nat. Genet. 49, 780–788 (2017).
Araf, S. et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 32, 1261–1265 (2018).
Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009).
Kalyana-Sundaram, S. et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia 14, 702–708 (2012).
Zhang, J. et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 26, 108–118 (2016).
Fang, L. T. et al. Establishing reference data and call sets for benchmarking cancer mutation detection using whole-genome sequencing. Preprint at bioRxiv https://doi.org/10.1101/625624 (2019).
Chen, X. et al. A multi-center cross-platform single-cell RNA sequencing reference dataset. Sci. Data 8, 39 (2021).
Chen, W. et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nature Biotechnol. https://www.nature.com/articles/s41587-020-00748-9 (2020).
Zhao, Y. et al. Whole genome and exome sequencing reference datasets from a multi-center and cross-platform benchmark study. Preprint at bioRxiv https://doi.org/10.1101/2021.02.27.433136 (2021).
Chen, L., Liu, P., Evans, T. C. & Ettwiller, L. M. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science 355, 752–756 (2017).
Costello, M. et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67 (2013).
Do, H. & Dobrovic, A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin. Chem. 61, 64–71 (2015).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ivanov, M. et al. Towards standardization of next-generation sequencing of FFPE samples for clinical oncology: intrinsic obstacles and possible solutions. J. Transl. Med. 15, 22 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. BFC: correcting Illumina sequencing errors. Bioinformatics 31, 2885–2887 (2015).
Freed, D., Pan, R. & Aldana, R. TNscope: accurate detection of somatic mutations with haplotype-based variant candidate detection and machine learning filtering. Preprint at bioRxiv https://doi.org/10.1101/250647 (2018).
Narzisi, G. et al. Lancet: genome-wide somatic variant calling using localized colored DeBruijn graphs. Commun. Biol. 1, 20 (2018).
Gargis, A. S. et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat. Biotechnol. 30, 1033–1036 (2012).
Chen, Y.-C. et al. Comprehensive assessment of somatic copy number variation calling using next-generation sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2021.02.18.431906 (2021).
Sahraeian, S. M. E., Fang, L. T., Mohiyuddin, M., Hong, H. & Xiao, W. Robust cancer mutation detection with deep learning models derived from tumor-normal sequencing data. Preprint at bioRxiv https://doi.org/10.1101/667261 (2019).
Tian, S. K. et al. Optimizing workflows and processing of cytologic samples for comprehensive analysis by next-generation sequencing: Memorial Sloan Kettering Cancer Center experience. Arch. Pathol. Lab. Med. 140, 1200–1205 (2016).
FastQC (Babraham Bioinformatics, accessed 2 July 2021); https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Picard (Broad Institute, accessed 2 July 2021); http://broadinstitute.github.io/picard/
Okonechnikov, K., Conesa, A. & García-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016).
Ewels, P. MultiQ. C. Aggregate results from bioinformatics analysis across many samples into a single report. Bioinformatics 32, 3047–3048 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank G. Sivakumar (Novartis) and S. Chacko (Center for Information Technology, National Institutes of Health (NIH)) for their assistance with data transfer, and J. Ye (Sentieon) for providing the Sentieon software package. We also thank D. Goldstein (Office of Technology and Science at the National Cancer Institute (NCI); L. Amundadottir (Division of Cancer Epidemiology and Genetics, NCI, NIH) for sponsorship and usage of the NIH Biowulf cluster; R. Phillip (Center for Devices and Radiological Health, US Food and Drug Administration) for advice on study design; and Seven Bridges for providing storage and computational support on the Cancer Genomic Cloud (CGC). The CGC has been funded in whole or in part with federal funds from the NCI, NIH (contract no. HHSN261201400008C and ID/IQ agreement no. 17×146 under contract no. HHSN261201500003I). Y. Zhao, J.L., T.-W.S., K.T., J.S., Y.K., A.R., B.T. and P.J. were supported by the Frederick National Laboratory for Cancer Research and through the NIH fund (NCI contract no. 75N910D00024). Research carried out in Charles Wang’s laboratory was partially supported by NIH grant no. S10OD019960 (to C.W.), the Ardmore Institute of Health (grant no. 2150141 to C.W.) and a Charles A. Sims gift. L.S. and Y. Zheng were supported by the National Key R&D Project of China (no. 2018YFE0201600), the National Natural Science Foundation of China (no. 31720103909) and Shanghai Municipal Science and Technology Major Project (no. 2017SHZDZX01). E.R. was supported by the European Union through the European Regional Development Fund (project no. 2014-2020.4.01.15-0012). J.N. and U.L. were supported by grants from the Swedish Research Council (no. 2017-00630/2019-01976). The work carried out at Palacky University was supported by grant no. LM2018132 from the Czech Ministry of Education, Youth and Sports. C.X. and S.T.S. were supported by the Intramural Research Program of the National Library of Medicine, NIH. This work also used the computational resources of the NIH Biowulf cluster (http://hpc.nih.gov). Original data were also backed up on servers provided by the Center for Biomedical Informatics and Information Technology, NCI. In addition, we thank the following individuals for their participation in working group discussions: M. Ashby, O. Aygun, X. Bian, P. Bushel, F. Campagne, T. Chen, H. Chuang, Y. Deng, D. Freed, P. Giresi, P. Gong, Y. Guo, C. Hatzis, S. Hester, J. Keats, E. Lee, Y. Li, S. Liang, T. McDianiel, J. Pandey, A. Pathak, T. Shi, J. Trent, M. Wang, X. Xu and C. Zhang. The following individuals from the University of Toledo Medical Center helped to troubleshoot or set up the FFPE protocol: A. Al-Agha, T. Cummins, C. Freeman, C. Nowak, A. Smigelski, J. Yeo and V. Kholodovych.
Author information
Authors and Affiliations
Contributions
The study was conceived and designed by W.X., L.S., E.D., S.L., L.M.S. and Z.T. Biosample preparation was performed by T.M.B., J.A.H., L.K., K.L., M.M. and J.C.W. NGS library preparation and sequencing were carried out by W.C., Z.C., J.D., A.G., T.H., K.I., H.J., E.J., R.K., B.S.M., S.K., Y.K., U.L., R.M., C.M., T.M., C. Miller, A.M., A.N., B.N., J.N., E.P., V.P., M. Polano, A.R., E.R., A.S, G.T.S., J.S., L.S., M.S., W.T., B.T., T.T., P.V., C.W., J.W. and Y. Zheng. Data analysis was performed by W.X., L.R., L.F., Y. Zhao, J.L., M.G., O.D.A., M.C., Q.C., X.C., Y.C., B.E., P.J., R.V.J., W.J., J.L., Z.L., X.L., C.L., D.M., U.M., C.M., C.N., B.N.P., M. Pirooznia, W.R., F.S., T.S., K.T., C.J.V., J.W., A.W., L.W., C.X., C.Y., G.Y., Y.Y. and B.Z. Data management was carried out by W.X., C.X., S.T.S., C.N. and D.M.. The manuscript was written by W.X., R.K., L.R., L.F., M.M., Y. Zhao and C.W. Project management was the responsibility of W.X., L.S., C.W., C.X. and H.H.
Corresponding authors
Ethics declarations
Competing interests
L.F. was an employees of Roche Sequencing Solutions Inc. L.K., K.L. and M.M. are employees of ATCC, which provided cell lines and derivative materials. E.J., O.D.A., T.T., A.M., A.N., A.G. and G.P.S. are employees of Illumina Inc. V.P. and M.S. are employees of Novartis Institutes for Biomedical Research. T.H., E.P. and R. Kalamegham are employees of Genentech (a member of the Roche group). Z.L. is an employee of Sentieon Inc. R.K. is an employee of Immuneering Corp. C.E.M. is a cofounder of Onegevity Health. All other authors claim no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Study design to capture “wet lab” factors affecting sequencing quality.
DNA was extracted from either fresh cells or FFPE processed cells (formalin fixation time of 1, 2, 6, or 24 hours). Both fresh DNA and FFPE DNA were profiled on WGS and WES platforms. For fresh DNA, six centers (Fudan University (FD), Illumina (IL), Novartis (NV), European Infrastructure for Translational Medicine (EA), National Cancer Institute (NC), and Loma Linda University (LL)) performed WGS and WES in parallel following manufacturer recommended protocols with limited deviation. Three of the six sequencing centers (FD, IL, and NV) generated library preparation in triplicate. For FFPE samples, each fixation time point had six blocks that were sequenced at two different centers (IL and GeneWiz (GZ)). Three library preparation protocols (TruSeq PCR-free, TruSeq-Nano, and Nextera Flex) were used with four different quantities of DNA input (1, 10, 100, and 250 ng) and sequenced by IL and LL. DNAs from HCC1395 and HCC1395BL were pooled at various ratios to create mixtures of 75%, 50%, 20%, 10%, and 5%. All libraries from these experiments were sequenced in triplicate on the HiSeq series by Genentech (GT). In addition, nine libraries using the TruSeq PCR-free preparation were run on a NovaSeq for WGS analysis by IL. Sample naming convention (example: WGS_FD_N_1): First field was used for sequencing study: Whole genome sequencing (WGS), Whole exome sequencing (WES), WGS on FFPE sample (FFG), WES on FFPE sample (FFX), WGS on library preparation protocol (LBP), WGS on tumor purity (SPP); Second field was used for sequencing centers, EA, FD, IL, LL, NC, NV, GT, and GZ or sequencing technologies, HiSeq (HS) and NovaSeq (NS); Third field was used for tumor (T) or normal (N); The last field was used for the number of repeats. *WGS performed only on Mixture (tumor purity) samples. ** WGS and WES performed only on FFPE samples.
Extended Data Fig. 2 Read mapping quality statistics.
(a) Percentage of reads mapped to target regions (SureSelect V6 + UTR) and G/C content for WES runs on fresh or FFPE DNA. (b) Read quality from three WGS library preparation kits (TruSeq PCRfree, TruSeq-Nano, and Nextera Flex) on fresh or FFPE DNA. (c) Distribution of GIV scores in WGS and WES runs. For detailed statistics regarding the boxplot, please refer to Supplementary Table 5.
Extended Data Fig. 3 Overall read quality distribution for all WES and WGS runs.
(a) Median insert fragment size of WES and WGS run on fresh and FFPE DNA. (b) G/C read content for WES and WGS runs. (c) Overall read redundancy for WES and WGS runs. Some outliers were observed in WGS on fresh DNA, which were from runs of TruSeq-Nano with 1 ng of DNA input. (d) Overall percentage of reads mapped to target regions for WES runs for fresh and FFPE DNA. For detailed statistics regarding the boxplot, please refer to Supplementary Table 6.
Extended Data Fig. 4 Mutation calling repeatability and O_Score distribution.
(a) Distribution of O_Score of three callers (MuTect2, Strelka2, and SomaticSniper) for twelve WGS and WES runs on BWA alignments. For detailed statistics regarding the boxplot, please refer to Supplementary Table 7. (b) “Tornado” plot of reproducibility between twelve WGS runs on the HiSeq series (2500, 4000, and X10) and nine WGS runs on the NovaSeq (S6000). SNVs/indels were called by Strelka2 on BWA alignments.
Extended Data Fig. 5 Source of variance in reproducibility measured by O_Score.
Actual by Predicted plot of WGS (a) and WES (b). A total of 8 variables (WGS) or 13 variables (WES), including 2-degree interactions, were included in the fixed effect linear model. 36 samples were used to derive statistics for both WES and WGS. The central blue line is the mean. The shaded region represents the 95% confidence interval.
Extended Data Fig. 6 Effect of post alignment processing on precision and recall of WES and WGS run on FFPE DNA.
(a) Precision and recall of mutation calls by Strelka2 on BWA alignments. A single library of FFPE DNA (FFX) and three libraries of fresh DNA (EA_1, FD_1, and NV_1) were run on a WES platform. Resulting reads were either processed by the BFC tool or by Trimmomatic. Processed FASTQ files were then aligned by BWA and called by Strelka2. Precision and recall were derived by matching calling results with the truth set. (b) Precision and recall of mutation calls by three callers, MuTect2 (blue), Strelka2 (green), and SomaticSniper (red), on BWA alignments without or with GATK post alignment process (indel realignment & BQSR).
Extended Data Fig. 7 Jaccard index scores to measure reproducibility of SNVs called by three callers.
Box plot of Jaccard scores of inter-center, intra-center, and overall pair of SNV call sets from two WGS or WES runs. SNVs were divided into three groups; Repeatable: SNVs defined in the truth set of the reference call set; Gray zone: SNVs not defined as “truth” in the reference call set; Non-Repeatable: SNVs were not in the reference call set. For detailed statistics regarding the boxplot, please refer to Supplementary Table 8.
Extended Data Fig. 8 Sources of variation in Jaccard index.
(a) Summary of factor effects. Twenty-five factors, including five original factors, ten 2-way interactions, and ten 3-way interactions were evaluated in the model. Both P values (derived from F-test) and their LogWorth (-log10 (P value)) are included in the summary plot. The factors are ordered by their LogWorth values. (b) Least square means of caller*pair_group*platform interaction. The height of the markers represents the adjusted least square means, and the bars represent confidence intervals of the means. (c) Least square means SNV_subset*pair_group*platform interaction. The height of the markers represents the adjusted least square means, and the bars represent confidence intervals of the means. 3168 samples were used to derive these statistics. (d) Student’s t-test for platform*pair_group interaction with SNV calls from three callers, MuTect2, Strelka2, and SomaticSniper. The left two panels compare Jaccard indices between intra-center and inter-center for WGS and WES, respectively. The right two panels compare Jaccard indices between WGS and WES for inter-center and intra-center pairs, respectively. Prob > |t| is the two-tailed test P value, and Prob>t is the one-tailed test P value.
Extended Data Fig. 9 WGS vs. WES platform-specific mutations and allele frequency calling accuracy.
Cumulative VAF plot of precision (a), recall (b), and F-Score (c) for three callers (MuTect2, Strelka2, and SomaticSniper) on WES and WGS runs.
Extended Data Fig. 10 Mutation allele frequency and coverage depth in WES and WGS sample.
Scatter plot of allele frequency and coverage depth by three callers, MuTect2, Strelka2, and SomaticSniper in one example WES sample (a) or WGS sample (b). (c) Boxplot of read depth on called mutations in WES or WGS. For detailed statistics regarding the boxplot, please refer to Supplementary Table 9.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1 and 2.
Supplementary Data
Supplementary Tables 1–10.
Rights and permissions
About this article
Cite this article
Xiao, W., Ren, L., Chen, Z. et al. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol 39, 1141–1150 (2021). https://doi.org/10.1038/s41587-021-00994-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00994-5
This article is cited by
-
Improving somatic exome sequencing performance by biological replicates
BMC Bioinformatics (2024)
-
Liquid biopsy in T-cell lymphoma: biomarker detection techniques and clinical application
Molecular Cancer (2024)
-
Computational immunogenomic approaches to predict response to cancer immunotherapies
Nature Reviews Clinical Oncology (2024)
-
Reference Materials for Improving Reliability of Multiomics Profiling
Phenomics (2024)
-
Reliable biological and multi-omics research through biometrology
Analytical and Bioanalytical Chemistry (2024)