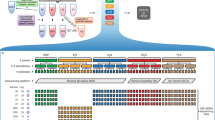
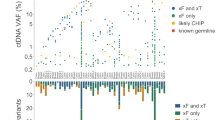
Abstract
Circulating tumor DNA (ctDNA) sequencing is being rapidly adopted in precision oncology, but the accuracy, sensitivity and reproducibility of ctDNA assays is poorly understood. Here we report the findings of a multi-site, cross-platform evaluation of the analytical performance of five industry-leading ctDNA assays. We evaluated each stage of the ctDNA sequencing workflow with simulations, synthetic DNA spike-in experiments and proficiency testing on standardized, cell-line-derived reference samples. Above 0.5% variant allele frequency, ctDNA mutations were detected with high sensitivity, precision and reproducibility by all five assays, whereas, below this limit, detection became unreliable and varied widely between assays, especially when input material was limited. Missed mutations (false negatives) were more common than erroneous candidates (false positives), indicating that the reliable sampling of rare ctDNA fragments is the key challenge for ctDNA assays. This comprehensive evaluation of the analytical performance of ctDNA assays serves to inform best practice guidelines and provides a resource for precision oncology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Descriptive data about individual ctDNA assays are provided in Supplementary Data 2. Descriptive data about individual variants, including their detection status in each ctDNA assay, are provided in variant classification tables within the Source Data Excel file. These tables were used to generate variant detection heat maps and other data plots. Raw sequencing data have been deposited to the National Center of Biotechnology Information Bioproject PRJNA677999. Variant calls generated by each assay vendor (in VCF format) and panel region files (in BED format) can be accessed at the following link: https://figshare.com/projects/SEQC2_Onco-panel_Sequencing_Working_Group_-_Liquid_Biopsy_Study/94523. Source data are provided with this paper.
Code availability
Variant call sets for each ctfDNA sequencing assay were generated by internal bioinformatics pipelines by each assay vendor. Although these pipelines are not open source, detailed descriptions and relevant software version numbers are provided in the Supplementary Methods. All data plots were generated using R (v3.5 or later) or GraphPad Prism (v8).
References
Leon, S. A., Shapiro, B., Sklaroff, D. M. & Yaros, M. J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650 (1977).
Stroun, M., Anker, P., Lyautey, J., Lederrey, C. & Maurice, P. A. Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol. 23, 707–712 (1987).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Aggarwal, C. et al. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat. Rev. Clin. Oncol. 18, 56–62 (2020).
Aravanis, A. M., Lee, M. & Klausner, R. D. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell 168, 571–574 (2017).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Volckmar, A.-L. et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer 57, 123–139 (2017).
Ross, M. G. et al. Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013).
Brannon, A. R. et al. Enhanced specificity of high sensitivity somatic variant profiling in cell-free DNA via paired normal sequencing: design, validation, and clinical experience of the MSK-ACCESS liquid biopsy assay. Preprint at bioRxiv https://doi.org/10.1101/2020.06.27.175471 (2020).
Clark, T. A. et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 20, 686–702 (2018).
Dawson, S.-J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 4, 136ra68 (2012).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K. W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011).
Klein, E. A. et al. Development of a comprehensive cell-free DNA (cfDNA) assay for early detection of multiple tumor types: the Circulating Cell-free Genome Atlas (CCGA) study. J. Clin. Oncol. 36, 12021–12021 (2018).
Miller, A. M. et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565, 654–658 (2019).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Murtaza, M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Odegaard, J. I. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 24, 3539 (2018).
Plagnol, V. et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS ONE 13, e0193802 (2018).
Kuderer, N. M. et al. Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol. 3, 996–998 (2017).
Stetson, D. et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis. Oncol. https://doi.org/10.1200/PO.18.00191 (2019).
Torga, G. & Pienta, K. J. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 4, 868–870 (2018).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Shiraishi, Y. et al. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 28, 1111–1125 (2018).
Bos, J. L. The ras gene family and human carcinogenesis. Mutat. Res. 195, 255–271 (1988).
Blackburn, J. et al. Use of synthetic DNA spike-in controls (sequins) for human genome sequencing. Nat. Protoc. 14, 2119–2151 (2019).
Deveson, I. W. et al. Chiral DNA sequences as commutable controls for clinical genomics. Nat. Commun. 10, 1342 (2019).
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013).
Jones, W. et al. A verified genomic reference sample for assessing performance of cancer panels detecting small variants of low allele frequency. Genome Biol. https://doi.org/10.1186/s13059-021-02316-z (2021).
Fu, G. K., Hu, J., Wang, P.-H. & Fodor, S. P. A. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc. Natl Acad. Sci. USA 108, 9026–9031 (2011).
Saito, T. & Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 10, e0118432 (2015).
Hardwick, S. A., Deveson, I. W. & Mercer, T. R. Reference standards for next-generation sequencing. Nat. Rev. Genet. 18, 473–484 (2017).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Sheridan, C. Investors keep the faith in cancer liquid biopsies. Nat. Biotechnol. 37, 972–974 (2019).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 10, e0140712 (2015).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Tie, J. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra92 (2016).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
Underhill, H. R. et al. Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Kim, Y.-W. et al. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp. Mol. Med. 51, 1–10 (2019).
Klega, K. et al. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis. Oncol. 2018, PO.17.00285 (2018).
Peng, H. et al. CNV detection from circulating tumor DNA in late stage non-small cell lung cancer patients. Genes (Basel) 10, 926 (2019).
Cai, Z. et al. Detection of microsatellite instability from circulating tumor DNA by targeted deep sequencing. J. Mol. Diagn. 22, 860–870 (2020).
Hu, Y. et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 24, 4437–4443 (2018).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Acknowledgements
All SEQC2 participants freely donated their time, reagents and computing resources for the completion and analysis of this project. We thank our expert colleague, S.-J. Dawson, for providing useful feedback during manuscript preparation. We acknowledge the following funding sources: NHMRC grants APP1108254 and APP1114016 (to T.R.M.), BAA grant HHSF223201510172C (to D.J.), Shanghai Municipal Science and Technology Major Project grant 2017SHZDZX01 (to L.S.), the National Natural Science Foundation of China grant 31720103909 (to L.S.), MRFF grant MRF1173594, Cancer Institute NSW Early Career Fellowship 2018/ECF013 and philanthropic support from the Kinghorn Foundation (to I.W.D.). The contents of the published materials are solely the responsibility of the administering institution, a participating institution or individual authors, and they do not reflect the views of any funding body listed above. This research includes contributions from, and was reviewed by, the FDA and the NIH. This work has been approved for publication by these agencies, but it does not necessarily reflect official agency policy. Certain commercial materials and equipment are identified to adequately specify experimental procedures. In no case does such identification imply recommendation or endorsement by the FDA or the NIH, nor does it imply that the items identified are necessarily the best available for the purpose.
Author information
Authors and Affiliations
Consortia
Contributions
W.T., D.J. and J.X. conceived the project. I.W.D., J.W., W.J., D.J., T.R.M. and J.X. devised the experiments. I.W.D. performed simulated experiments. B.S.M., J.B., I.S., A.B., D.C., J.C., M.H., N.M., P.M., R.S., D.S., L.S., P.S., H.T., L.T., D.T., H.A., H.B., B.B., D.D., A.G., S.G., K.H., C.M., A.R., P.R., R.R., R.S., M.S., P.S., M.S., V.T. and S.V. performed and/or coordinated laboratory experiments. I.W.D. and B.G. performed data analysis. I.W.D. and T.R.M. prepared the figures. I.W.D., D.J., T.R.M. and J.X. prepared the manuscript, with support from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The Garvan Institute of Medical Research has filed patent applications on synthetic controls for genomics.
Additional information
Peer review information Nature Biotechnology thanks Michael Berger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Supplementary Tables 1–6 and Supplementary Methods
Supplementary Data 1
Description of synthetic ‘sequin’ mutations
Supplementary Data 2
Summary of variant detection across ctDNA assays
Supplementary Data 3
Extraction yields from synthetic plasma
Source data
Main Figures
Descriptive data about individual variants, including their detection status in each ctDNA assay, are provided in variant classification tables within the Source Data Excel file.
Rights and permissions
About this article
Cite this article
Deveson, I.W., Gong, B., Lai, K. et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol 39, 1115–1128 (2021). https://doi.org/10.1038/s41587-021-00857-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00857-z
This article is cited by
-
Analytical evaluation of circulating tumor DNA sequencing assays
Scientific Reports (2024)
-
Reference Materials for Improving Reliability of Multiomics Profiling
Phenomics (2024)
-
Reliable biological and multi-omics research through biometrology
Analytical and Bioanalytical Chemistry (2024)
-
Blood-based liquid biopsy: insights into early detection, prediction, and treatment monitoring of bladder cancer
Cellular & Molecular Biology Letters (2023)
-
Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma
Biomarker Research (2023)