Abstract
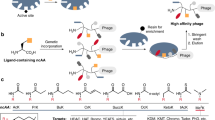
Molecules that covalently bind macromolecular targets have found widespread applications as activity-based probes and as irreversibly binding drugs. However, the general reactivity of the electrophiles needed for covalent bond formation makes control of selectivity difficult. There is currently no rapid, unbiased screening method to identify new classes of covalent inhibitors from highly diverse pools of candidate molecules. Here we describe a phage display method to directly screen for ligands that bind to protein targets through covalent bond formation. This approach makes use of a reactive linker to form cyclic peptides on the phage surface while simultaneously introducing an electrophilic ‘warhead’ to covalently react with a nucleophile on the target. Using this approach, we identified cyclic peptides that irreversibly inhibited a cysteine protease and a serine hydrolase with nanomolar potency and exceptional specificity. This approach should enable rapid, unbiased screening to identify new classes of highly selective covalent inhibitors for diverse molecular targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data presented in this manuscript are available from the corresponding author upon reasonable request. The TEV protease-expressing plasmid sequence is available at GenBank with accession number MN480436. Characterization data for cyclization linker and probes are available in the Supplementary Information. Source data are provided with this paper.
References
Singh, J., Petter, R. C., Baillie, T. A. & Whitty, A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011).
Ghosh, A. K., Samanta, I., Mondal, A. & Liu, W. R. Covalent inhibition in drug discovery. ChemMedChem 14, 889–906 (2019).
Cravatt, B. F., Wright, A. T. & Kozarich, J. W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 (2008).
Sanman, L. E. & Bogyo, M. Activity-based profiling of proteases. Annu. Rev. Biochem. 83, 249–273 (2014).
Cohen, M. S., Zhang, C., Shokat, K. M. & Taunton, J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science 308, 1318–1321 (2005).
Smith, G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Matthews, D. J. & Wells, J. A. Substrate phage: selection of protease substrates by monovalent phage display. Science 260, 1113–1117 (1993).
Heinis, C., Rutherford, T., Freund, S. & Winter, G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 5, 502–507 (2009).
Chen, S., Bertoldo, D., Angelini, A., Pojer, F. & Heinis, C. Peptide ligands stabilized by small molecules. Angew. Chem. Int. Ed. Engl. 53, 1602–1606 (2014).
Jafari, M. R. et al. Discovery of light-responsive ligands through screening of a light-responsive genetically encoded library. ACS Chem. Biol. 9, 443–450 (2014).
Ng, S. & Derda, R. Phage-displayed macrocyclic glycopeptide libraries. Org. Biomol. Chem. 14, 5539–5545 (2016).
Cardote, T. A. & Ciulli, A. Cyclic and macrocyclic peptides as chemical tools to recognise protein surfaces and probe protein–protein interactions. ChemMedChem 11, 787–794 (2016).
Cesaratto, F., Burrone, O. R. & Petris, G. Tobacco etch virus protease: a shortcut across biotechnologies. J. Biotechnol. 231, 239–249 (2016).
Lentz, C. S. et al. Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP). Nat. Chem. Biol. 14, 609–617 (2018).
Assem, N., Ferreira, D. J., Wolan, D. W. & Dawson, P. E. Acetone-linked peptides: a convergent approach for peptide macrocyclization and labeling. Angew. Chem. Int. Ed. Engl. 54, 8665–8668 (2015).
Larsen, D. et al. Exceptionally rapid oxime and hydrazone formation promoted by catalytic amine buffers with low toxicity. Chem. Sci. 9, 5252–5259 (2018).
Palmer, J. T., Rasnick, D., Klaus, J. L. & Bromme, D. Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem. 38, 3193–3196 (1995).
Oleksyszyn, J., Boduszek, B., Kam, C. M. & Powers, J. C. Novel amidine-containing peptidyl phosphonates as irreversible inhibitors for blood coagulation and related serine proteases. J. Med. Chem. 37, 226–231 (1994).
Timmerman, P., Beld, J., Puijk, W. C. & Meloen, R. H. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. Chembiochem 6, 821–824 (2005).
Kather, I., Bippes, C. A. & Schmid, F. X. A stable disulfide-free gene-3-protein of phage fd generated by in vitro evolution. J. Mol. Biol. 354, 666–678 (2005).
Chen, S., Morales-Sanfrutos, J., Angelini, A., Cutting, B. & Heinis, C. Structurally diverse cyclisation linkers impose different backbone conformations in bicyclic peptides. Chembiochem 13, 1032–1038 (2012).
Hornsby, M. et al. A high through-put platform for recombinant antibodies to folded proteins. Mol. Cell. Proteom. 14, 2833–2847 (2015).
Raran-Kurussi, S., Cherry, S., Zhang, D. & Waugh, D. S. Removal of affinity tags with TEV protease. Methods Mol. Biol. 1586, 221–230 (2017).
Rebollo, I. R., Angelini, A. & Heinis, C. Phage display libraries of differently sized bicyclic peptides. MedChemComm 4, 145–150 (2013).
Nazif, T. & Bogyo, M. Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc. Natl Acad. Sci. USA 98, 2967–2972 (2001).
Albrow, V. E. et al. Development of small molecule inhibitors and probes of human SUMO deconjugating proteases. Chem. Biol. 18, 722–732 (2011).
Hemelaar, J. et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell Biol. 24, 84–95 (2004).
Choe, Y. et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 281, 12824–12832 (2006).
Bromme, D., Klaus, J. L., Okamoto, K., Rasnick, D. & Palmer, J. T. Peptidyl vinyl sulphones: a new class of potent and selective cysteine protease inhibitors: S2P2 specificity of human cathepsin O2 in comparison with cathepsins S and L. Biochem. J. 315, 85–89 (1996).
Sanman, L. E., van der Linden, W. A., Verdoes, M. & Bogyo, M. Bifunctional probes of cathepsin protease activity and pH reveal alterations in endolysosomal pH during bacterial infection. Cell Chem. Biol. 23, 793–804 (2016).
Verdoes, M. et al. Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J. Am. Chem. Soc. 135, 14726–14730 (2013).
Salomon-Ferrer, R., Case, D. A. & Walker, R. C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 3, 198–210 (2013).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006).
Phan, J. et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277, 50564–50572 (2002).
Chen, L., Keller, L. J., Cordasco, E., Bogyo, M. & Lentz, C. S. Fluorescent triazole urea activity-based probes for the single-cell phenotypic characterization of Staphylococcus aureus. Angew. Chem. Int. Ed. Engl. 58, 5643–5647 (2019).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Hacker, S. M. et al. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 (2017).
Kapust, R. B. et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001).
Shen, A. et al. Simplified, enhanced protein purification using an inducible, autoprocessing enzyme tag. PLoS ONE 4, e8119 (2009).
Fellner, M. et al. Structural basis for the inhibitor and substrate specificity of the unique Fph serine hydrolases of Staphylococcus aureus. ACS Infect. Dis. 6, 2771–2782 (2020).
Bellotto, S., Chen, S., Rentero Rebollo, I., Wegner, H. A. & Heinis, C. Phage selection of photoswitchable peptide ligands. J. Am. Chem. Soc. 136, 5880–5883 (2014).
Chen, S. et al. Improving binding affinity and stability of peptide ligands by substituting glycines with d-amino acids. Chembiochem 14, 1316–1322 (2013).
Mikolajczyk, J. et al. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 282, 26217–26224 (2007).
Li, H. et al. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530, 233–236 (2016).
Chen, S. et al. Bicyclic peptide ligands pulled out of cysteine-rich peptide libraries. J. Am. Chem. Soc. 135, 6562–6569 (2013).
Lentz, C. S. et al. Design of selective substrates and activity-based probes for hydrolase important for pathogenesis 1 (HIP1) from Mycobacterium tuberculosis. ACS Infect. Dis. 2, 807–815 (2016).
Chen, S. et al. Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides. Nat. Chem. 6, 1009–1016 (2014).
Acknowledgements
We thank the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University for providing technical assistance with mass spectrometry. We also thank D. Waugh (National Cancer Institute) for providing the TEV expression construct, pDZ2087, and C. Heinis (EPFL) for providing the phage library. This work was supported by Swiss National Science Foundation Postdoc Mobility fellowship P2ELP3_155323 P300PB_164725 (to S.C.) and by funding from National Institutes of Health grants R01 EB026285 and R01 EB026285 02S1 (to M.B.).
Author information
Authors and Affiliations
Contributions
M.B. and S.C. conceived the project and designed the experiments. S.C., S. Lovell, S. Lee and M.F. performed the experiments and analyzed the data. M.B. and S.C. wrote the manuscript with input from all authors. M.B. and P.D.M. obtained funding for the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Strategy for the synthesis of DCA-VS, derivatizing a vinyl sulfone cysteine reactive warhead with 1,3-Dichloroacetone.
The cysteine protease reactive vinyl sulfone warhead was efficiently conjugated to the 1,2-dichloroacetone linker through oxime-ketone reaction.
Extended Data Fig. 2 Strategy for the synthesis of DCA-DPP, derivatizing a diphenylphosphonate serine reactive warhead with 1,3-Dichloroacetone.
a, Acetic Anhydride, paraformaldehyde, triphenyl phosphite, acetic acid, 5 h, 120 °C, 46 % b, i. HBr in acetic acid ii. (Boc-aminooxy)acetic acid, DCC, DIPEA, DMF, RT, O/N, 67 % c, i. TFA in DCM, RT, 1 h ii. Dichloroacetone, DMF, RT, O/N, 72 %.
Extended Data Fig. 3 Strategy for synthesis of the Fmoc-Gln-vinyl sulfone warhead.
The carboxyl side chain of P1 glutamine was deprotected and reacted with chlorotrityl resin. The derived resin can be used for directly synthesizing linear ABPs bearing a glutamine-VS motif at the C-terminus.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14.
Supplementary Video 1
Molecular dynamic simulation of TEV13 complex with TEV protease.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed gels.
Rights and permissions
About this article
Cite this article
Chen, S., Lovell, S., Lee, S. et al. Identification of highly selective covalent inhibitors by phage display. Nat Biotechnol 39, 490–498 (2021). https://doi.org/10.1038/s41587-020-0733-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0733-7
This article is cited by
-
A single-component, light-assisted uncaging switch for endoproteolytic release
Nature Chemical Biology (2024)
-
High-throughput fabrication of antimicrobial phage microgels and example applications in food decontamination
Nature Protocols (2024)
-
Identification of photocrosslinking peptide ligands by mRNA display
Communications Chemistry (2023)
-
Discovering covalent inhibitors of protein–protein interactions from trillions of sulfur(VI) fluoride exchange-modified oligonucleotides
Nature Chemistry (2023)
-
A simple method for developing lysine targeted covalent protein reagents
Nature Communications (2023)