Abstract
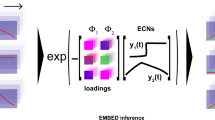
The translational power of human microbiome studies is limited by high interindividual variation. We describe a dimensionality reduction tool, compositional tensor factorization (CTF), that incorporates information from the same host across multiple samples to reveal patterns driving differences in microbial composition across phenotypes. CTF identifies robust patterns in sparse compositional datasets, allowing for the detection of microbial changes associated with specific phenotypes that are reproducible across datasets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The sequences and biome tables for the IBD, ECAM, DIABIMMUNE and AGP datasets can be found on Qiita (http://qiita.microbio.me) under study IDs 1629, 10249, 11884 and 10317 and at EBI or BioProject under ERP020401, ERP016173, PRJNA290381 and ERP012803.
Code availability
The CTF codebase named Gemelli is a fully unit tested open-source python package, and is installable through pip or conda. Additionally, CTF is wrapped in a QIIME2 plugin: https://github.com/biocore/gemelli; all the code and analyses are available in the ‘Code Ocean’ capsule: https://doi.org/10.24433/CO.5938114.v1.
References
Gibson, T. E. & Gerber, G. K. Robust and scalable models of microbiome dynamics. In Proceedings of the 35th International Conference on Machine Learning 80 (eds Dy, J. et al.) 1763–1772 (PMLR, 2018).
Shenhav, L. et al. Modeling the temporal dynamics of the gut microbial community in adults and infants. PLoS Comput. Biol. 15, e1006960 (2019).
Äijö, T., Müller, C. L. & Bonneau, R. Temporal probabilistic modeling of bacterial compositions derived from 16S rRNA sequencing. Bioinformatics 34, 372–380 (2018).
Silverman, J. D., Durand, H. K., Bloom, R. J., Mukherjee, S. & David, L. A. Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome 6, 202 (2018).
Martino, C. et al. A novel sparse compositional technique reveals microbial perturbations. mSystems 4, e00016–e00019 (2019).
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V. & Egozcue, J. J. Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8, 2224 (2017).
Morton, J. T. et al. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 10, 2719 (2019).
Halfvarson, J. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2, 17004 (2017).
Jaccard, P. The distribution of the flora in the alpine zone. 1. New Phytol. 11, 37–50 (1912).
Bray, J. R. & Curtis, J. T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957).
Aitchison, J. Principal component analysis of compositional data. Biometrika 70, 57–65 (1983).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
McDonald, D. et al. Striped UniFrac: enabling microbiome analysis at unprecedented scale. Nat. Methods 15, 847–848 (2018).
Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82–343ra82 (2016).
Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016).
McDonald, D. et al. American Gut: an open platform for citizen science microbiome research. mSystems 3, e00031–18 (2018).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Keshavan, R. H., Montanari, A. & Oh, S. Low-rank matrix completion with noisy observations: a quantitative comparison. In Proc. 2009 47th Annual Allerton Conference on Communication, Control, and Computing 1216–1222 (Curran Associates, 2009).
Lek-Heng Lim. Singular values and eigenvalues of tensors: a variational approach. In Proc. 1st IEEE International Workshop on Computational Advances in Multi-Sensor Adaptive Processing 129–132 (Curran Associates, 2005).
Anandkumar, A., Ge, R. & Janzamin, M. Guaranteed non-orthogonal tensor decomposition via alternating rank-1 updates. Preprint at arXiv http://arxiv.org/abs/1402.5180 (2014).
Jain, P. & Oh, S. Provable tensor factorization with missing data. Adv. Neural Inf. Process. Syst. 27 (eds Ghahramani, Z. et al.) 1431–1439 (Curran Associates, 2014).
Aitchison, J. & Ho, C. H. The multivariate Poisson-log normal distribution. Biometrika 76, 643–653 (1989).
Amir, A. et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–16 (2017).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Janssen, S. et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 3, e00021–18 (2018).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Gonzalez, A. et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods 551, 457 (2018).
Acknowledgements
This work was partially supported by the EMCH fund for human microbiome studies, the Norwegian Institute of Public Health 2019-0350 (R.K.), the Emerald Foundation 3022 (R.K.), National Institutes of Health (NIH) Pioneer award grant no. 1DP1AT010885 (R.K.), National Institute of Justice grant no. 2016-DN-BX-4194 (R.K.), San Diego Digestive Diseases Research Center NIDDK grant no. 1P30DK120515 (R.K.) and Janssen Pharmaceuticals grant no. 20175015 (R.K.). C.A.M. was funded by the NIDCR (grant no. 1F31DE028478-01). E.H. and L.S were partially supported by the National Science Foundation (grant no. 1705197) and by NIH grant no. 1R56MD013312. E.H. was also partially supported by grants no. NIH/NHGRI HG010505-02, NIH 1R01MH115979, NIH 5R25GM112625 and NIH 5UL1TR001881. M.G.D.-B. was funded in part by the C&D Research Fund.
Author information
Authors and Affiliations
Contributions
C.M., L.S. and R.K. conceived, initiated and coordinated the project. C.M., L.S., D.M. and Y.V.-B. coordinated, compiled and performed analysis. C.M., C.A.M. and G.A. wrote the code for CTF. C.M., L.S. and C.A.M. wrote the manuscript. J.T.M, A.D.S. and M.G.D.-B. provided essential discussion and advice. E.H. and R.K. supervised the project. All authors discussed the experiments and results, read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion, Figs. 1–6, Tables 1–3 and References.
Rights and permissions
About this article
Cite this article
Martino, C., Shenhav, L., Marotz, C.A. et al. Context-aware dimensionality reduction deconvolutes gut microbial community dynamics. Nat Biotechnol 39, 165–168 (2021). https://doi.org/10.1038/s41587-020-0660-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0660-7
This article is cited by
-
Revealing the role of the gut microbiota in enhancing targeted therapy efficacy for lung adenocarcinoma
Experimental Hematology & Oncology (2024)
-
Analyzing postprandial metabolomics data using multiway models: a simulation study
BMC Bioinformatics (2024)
-
A conserved interdomain microbial network underpins cadaver decomposition despite environmental variables
Nature Microbiology (2024)
-
Microbiome response in an urban river system is dominated by seasonality over wastewater treatment upgrades
Environmental Microbiome (2023)
-
Synthetic living materials in cancer biology
Nature Reviews Bioengineering (2023)