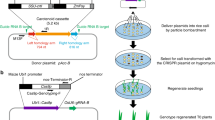
Abstract
CRISPR–Cas9 methods have been applied to generate random insertions and deletions, large deletions, targeted insertions or replacements of short sequences, and precise base changes in plants1,2,3,4,5,6,7. However, versatile methods for targeted insertion or replacement of long sequences and genes, which are needed for functional genomics studies and trait improvement in crops, are few and largely depend on the use of selection markers8,9,10,11. Building on methods developed in mammalian cells12, we used chemically modified donor DNA and CRISPR–Cas9 to insert sequences of up to 2,049 base pairs (bp), including enhancers and promoters, into the rice genome at an efficiency of 25%. We also report a method for gene replacement that relies on homology-directed repair, chemically modified donor DNA and the presence of tandem repeats at target sites, achieving replacement with up to 130-bp sequences at 6.1% efficiency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of the present study are available in the article and its supplementary figures and tables, or from the corresponding author upon request. For sequence data, rice LOC_Os IDs listed in Supplementary Table 1 are available on the Rice Genome Annotation Project site (http://rice.plantbiology.msu.edu/). The deep sequencing data were deposited with the National Center for Biotechnology Information BioProject database under the accession code PRJNA608130. Source data are provided with this paper.
Code availability
Custom script for analyzing NGS data is available at https://github.com/zhulab-ge/knockin. Source data are provided with this paper.
References
Zhang, H., Zhang, J., Lang, Z., Botella, J. R. & Zhu, J.-K. Genome editing—principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36, 291–309 (2017).
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H. & Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914 (2014).
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Lu, Y. & Zhu, J. K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 System. Mol. Plant 10, 523–525 (2017).
Lin, Q. et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020).
Mao, Y., Botella, J. R., Liu, Y. & Zhu, J.-K. Gene editing in plants: progress and challenges. Natl Sci. Rev. 6, 421–437 (2019).
Sun, Y. et al. Engineering herbicide-resistant rice plants through crispr/cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9, 628–631 (2016).
Sauer, N. J. et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170, 1917–1928 (2016).
Li, J. et al. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2, 16139 (2016).
Wang, M. et al. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 10, 1007–1010 (2017).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14 (2005).
Sugio, T., Satoh, J., Matsuura, H., Shinmyo, A. & Kato, K. The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J. Biosci. Bioeng. 105, 300–302 (2008).
Ren, Z. H. et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146 (2005).
Uga, Y. et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102 (2013).
Shi, H., Lee, B. H., Wu, S. J. & Zhu, J. K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85 (2003).
Ikeda, A. et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010 (2001).
Jobling, S. A. & Gehrke, L. Enhanced translation of chimaeric messenger RNAs containing a plant viral untranslated leader sequence. Nature 325, 622–625 (1987).
Rouached, H., Secco, D., Arpat, B. & Poirier, Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 11, 19 (2011).
Sahoo, D. K., Sarkar, S., Raha, S., Maiti, I. B. & Dey, N. Comparative analysis of synthetic DNA promoters for high-level gene expression in plants. Planta 240, 855–875 (2014).
Feng, Z. et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 4632–4637 (2014).
Li, X. M. et al. Natural alleles of a proteasome alpha2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833 (2015).
Hu, B. et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838 (2015).
Lu, Y. et al. Genome-wide targeted mutagenesis in rice using CRISPR/Cas9 system. Mol. Plant 10, 1242–1245 (2017).
Liu, H. et al. CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532 (2017).
Nishimura, A., Aichi, I. & Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802 (2006).
Liu, W. et al. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 8, 1431–1433 (2015).
Acknowledgements
This work was financially supported by the Chinese Academy of Sciences, including the CAS Strategic Priority Research Program grant no. XDB27040101 to J.-K.Z., and by the Major Project of China on New Varieties of GMO Cultivation (grant no. 2019ZX08010-003 to F.L.).
Author information
Authors and Affiliations
Contributions
Y.L. and J.-K.Z. designed the experiments. M.C., Y.T., R.S., J.D., F.L. and T.Z. performed the rice transformations. Y.L., Y.T., R.S., Q.Y., M.C., M.W., J.D., T.Z. and M.L. performed all the other experiments. Y.L. and J.-K.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Tables 1–6.
Source data
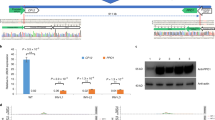
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Unprocessed gels and western blots.
Rights and permissions
About this article
Cite this article
Lu, Y., Tian, Y., Shen, R. et al. Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol 38, 1402–1407 (2020). https://doi.org/10.1038/s41587-020-0581-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0581-5
This article is cited by
-
Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis
BMC Biology (2024)
-
Precise integration of large DNA sequences in plant genomes using PrimeRoot editors
Nature Biotechnology (2024)
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Cost-effective multiplex PCR assay for simultaneous detection of bacterial leaf blight, blast and brown planthopper resistance genes in rice
Journal of Plant Biochemistry and Biotechnology (2024)
-
Breeding rice for yield improvement through CRISPR/Cas9 genome editing method: current technologies and examples
Physiology and Molecular Biology of Plants (2024)