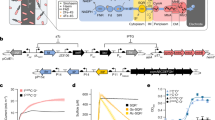
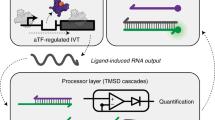
Abstract
Lack of access to safe drinking water is a global problem, and methods to reliably and easily detect contaminants could be transformative. We report the development of a cell-free in vitro transcription system that uses RNA Output Sensors Activated by Ligand Induction (ROSALIND) to detect contaminants in water. A combination of highly processive RNA polymerases, allosteric protein transcription factors and synthetic DNA transcription templates regulates the synthesis of a fluorescence-activating RNA aptamer. The presence of a target contaminant induces the transcription of the aptamer, and a fluorescent signal is produced. We apply ROSALIND to detect a range of water contaminants, including antibiotics, small molecules and metals. We also show that adding RNA circuitry can invert responses, reduce crosstalk and improve sensitivity without protein engineering. The ROSALIND system can be freeze-dried for easy storage and distribution, and we apply it in the field to test municipal water supplies, demonstrating its potential use for monitoring water quality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data presented in this manuscript are available as supplementary data files. All plasmids used in this manuscript are available in Addgene with the identifiers 140371–140399. Source data are provided with this paper.
References
French, K. E. Harnessing synthetic biology for sustainable development. Nat. Sustain. 2, 250–252 (2019).
Slomovic, S., Pardee, K. & Collins, J. J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl Acad. Sci. USA 112, 14429–14435 (2015).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Takahashi, M. K. et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 9, 3347 (2018).
Vorosmarty, C. J. et al. Global threats to human water security and river biodiversity. Nature 467, 555–561 (2010).
van der Meer, J. R. & Belkin, S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol. 8, 511–522 (2010).
McNerney, M. P. et al. Active analyte import improves the dynamic range and sensitivity of a vitamin B12 biosensor. ACS Synth. Biol. 9, 402–411 (2020).
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310 (2017).
Siegal-Gaskins, D. et al. Gene circuit performance characterization and resource usage in a cell-free ‘breadboard’. ACS Synth. Biol. 3, 416–425 (2014).
Alam, K. K. et al. A fluorescent split aptamer for visualizing RNA–RNA assembly in vivo. ACS Synth. Biol. 6, 1710–1721 (2017).
Zhang, Z. et al. Single-molecule tracking of the transcription cycle by sub-second RNA detection. eLife 3, e01775 (2014).
Pedelacq, J. D. et al. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Cuthbertson, L. & Nodwell, J. R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475 (2013).
Dunn, J. J. & Studier, F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166, 477–535 (1983).
Fernandez-Lopez, R. et al. Transcription factor-based biosensors enlightened by the analyte. Front. Microbiol. 6, 648 (2015).
Baquero, F., Martinez, J. L. & Canton, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265 (2008).
Zhang, W. et al. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl. Environ. Microbiol. 72, 2573–2580 (2006).
Deochand, D. K. & Grove, A. MarR family transcription factors: dynamic variations on a common scaffold. Crit. Rev. Biochem. Mol. Biol. 52, 595–613 (2017).
Zhu, T. et al. Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab. Eng. 19, 69–78 (2013).
Noguchi, N. et al. Regulation of transcription of the mph(A) gene for macrolide 2′-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J. Bacteriol. 182, 5052–5058 (2000).
Richardson, S. D. & Ternes, T. A. Water analysis: emerging contaminants and current issues. Anal. Chem. 90, 398–428 (2018).
Kasey, C. M. et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology. ACS Synth. Biol. 7, 227–239 (2018).
Carballa, M. et al. Behavior of pharmaceuticals, cosmetics and hormones in a sewage treatment plant. Water Res. 38, 2918–2926 (2004).
Hiromoto, T. et al. Characterization of MobR, the 3-hydroxybenzoate-responsive transcriptional regulator for the 3-hydroxybenzoate hydroxylase gene of Comamonas testosteroni KH122-3s. J. Mol. Biol. 364, 863–877 (2006).
Kim, M. et al. Widely used benzalkonium chloride disinfectants can promote antibiotic resistance. Appl. Microbiol. 84, e01201-18. (2018).
Grkovic, S. et al. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273, 18665–18673 (1998).
Lu, W. J. et al. Enantiomers of naringenin as pleiotropic, stereoselective inhibitors of cytochrome P450 isoforms. Chirality 23, 891–896 (2011).
Rogers, J. K. et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 43, 7648–7660 (2015).
Liu, X. et al. De novo design of programmable inducible promoters. Nucleic Acids Res. 47, 10452–10463 (2019).
Juraschek, S. P. et al. Gout, urate-lowering therapy, and uric acid levels among adults in the United States. Arthrit. Care Res. 67, 588–592 (2015).
Wilkinson, S. & Grove, A. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 279, 51442–51450 (2004).
Abraham, J., Dowling, K. & Florentine, S. Risk of post-fire metal mobilization into surface water resources: a review. Sci. Total Environ. 599, 1740–1755 (2017).
Hanna-Attisha, M. et al. Elevated blood lead levels in children associated with the Flint drinking water crisis: a spatial analysis of risk and public health response. Am. J. Public Health 106, 283–290 (2016).
Calle, G. R. et al. Enhanced copper release from pipes by alternating stagnation and flow events. Environ. Sci. Technol. 41, 7430–7436 (2007).
VanZile, M. L., Chen, X. & Giedroc, D. P. Allosteric negative regulation of smt O/P binding of the zinc sensor, SmtB, by metal ions: a coupled equilibrium analysis. Biochemistry 41, 9776–9786 (2002).
Chang, F. M. et al. Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 289, 19204–19217 (2014).
Busenlehner, L. S. et al. Spectroscopic properties of the metalloregulatory Cd(II) and Pb(II) sites of S. aureus pI258 CadC. Biochemistry 40, 4426–4436 (2001).
Lloyd, J. et al. Dynamic control of aptamer–ligand activity using strand displacement reactions. ACS Synth. Biol. 7, 30–37 (2018).
Reyes-Caballero, H. et al. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J. Mol. Biol. 403, 197–216 (2010).
Danner, M. C. et al. Antibiotic pollution in surface fresh waters: occurrence and effects. Sci. Total Environ. 664, 793–804 (2019).
Steber, M. et al. Mechanistic basis for RNA aptamer-based induction of TetR. ChemBioChem 12, 2608–2614 (2011).
Pardee, K. et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014).
Haklay, M. A. W. P. Openstreetmap: UserGenerated street maps. IEEE Pervas. Comput. 7, 12–18 (2008).
Vega, A. S., Lizama, K. & Pastén, P. A. in Water Policy in Chile (ed. Donoso, G.) 25–51 (Springer International Publishing, 2018).
Chen, X. et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 37, 1287–1293 (2019).
Ellefson, J. W., Ledbetter, M. P. & Ellington, A. D. Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat. Chem. Biol. 14, 361–367 (2018).
McNerney, M. P. et al. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 5, eaax4473 (2019).
Capdevila, D. A. et al. Tuning site-specific dynamics to drive allosteric activation in a pneumococcal zinc uptake regulator. eLife 7, e37268 (2018).
Acknowledgements
We thank A. Thompson (Northwestern University) for managing the experimental reagents and equipment used in this study; Z. Yang (Northwestern University) for assistance with FAAS measurements; N. Donghia (Wyss Institute) for helpful discussions on lyophilization of cell-free reactions; S. Pshenychny (Recombinant Protein Production Core at Northwestern University) for assistance in protein purification; J. Webster, R. Gruenberg, J. Machado and E. Plasencia-Campos (CSU, Chico) for assisting with field work; and J. Bussan, F. Lantz and B. Golenia (Northwestern University Research Shop) for assistance in the development of the handheld illuminator. J.K.J. and M.S.V. were supported in part by Northwestern University’s Graduate School Cluster in Biotechnology, System, and Synthetic Biology, which is affiliated with the Biotechnology Training Program. P.R.C. was supported in part by the National Science Foundation (NSF) Synthetic Biology REU (grant no. DBI-1757973). This work was also supported by funding from the Pew Charitable Trusts (to D.A.C.), the National Institutes of Health (grant no. R35 GM118157 to D.P.G.), CONICYT/FONDECYT (grant no. 1161337 to P.A.P.), ANID/FONDAP (grant no. 15110020 to P.A.P.), NSF CAREER (grant no. 1452441 to J.B.L.), NSF MCB RAPID (grant no. 1929912 to J.B.L., J.-F.G. and S.J.M.), support from the Crown Family Center for Jewish and Israel Studies at Northwestern University (to J.B.L.) and Searle Funds at the Chicago Community Trust (to J.B.L.).
Author information
Authors and Affiliations
Contributions
J.K.J., K.K.A., J.J.C. and J.B.L. conceived the project. J.K.J., K.K.A., M.S.V. and J.B.L. curated the data. J.K.J., K.K.A. and J.B.L. carried out the formal analysis. J.B.L., J.-F.G., S.J.M., D.P.G. and P.A.P. acquired the funds. J.K.J., K.K.A., M.S.V., M.D., P.R.C., J.W.L. and J.-F.G. carried out the investigations. J.K.J., K.K.A., M.S.V., J.W.L., P.Q.N., D.A.C., M.D., S.J.M., J.-F.G. and J.B.L. devised the methodology. J.K.J., K.K.A. and J.B.L. administered the project. D.A.C., D.P.G., P.A.P. and S.J.M. acquired the resources. J.K.J., K.K.A., J.J.C. and J.B.L. supervised the project. J.K.J., K.K.A., M.S.V., M.D., P.R.C. and J.-F.G. validated the project. J.K.J., K.K.A. and J.B.L. visualized the project. J.K.J., K.K.A. and J.B.L. wrote the original draft. J.K.J., K.K.A., M.S.V., M.D., P.R.C., J.W.L., P.Q.N., D.A.C., P.A.P., S.J.M., J.-F.G., D.P.G., J.J.C. and J.B.L. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
K.K.A., J.K.J. and J.B.L. have submitted a US provisional patent application (no. 62/758,242) relating to regulated IVT reactions. K.K.A., J.K.J., M.S.V., P.R.C., J.W.L., J.J.C. and J.B.L. have submitted a US provisional patent application (no. 62/838,852) relating to the preservation and stabilization of IVT reactions. K.K.A. and J.B.L. are founders and have a financial interest in Stemloop, Inc. The latter interests are reviewed and managed by Northwestern University in accordance with their conflict-of-interest policies. All other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Homemade transcription reactions can be optimized to perform as well as commercial kits.
Homemade in vitro transcription reactions were compared to a commercially available high yield transcription kit (NEB HiScribe™ T7 Quick High Yield RNA Synthesis Kit). 25 nM DNA encoding T7–3WJdB-T was added to each reaction in a total reaction volume of 20 µL. Over the course of 1 hour, the data show similar fluorescence activation. However, the homemade reaction begins to saturate after 1 hour, likely due to the exhaustion of NTPs in the homemade reaction (11.4 mM) when compared to the commercial kit (40 mM). All data shown for n = 3 independent biological replicates as lines with raw fluorescence values standardized to MEF (µM FITC). Shading indicates the average value of 3 independent biological replicates ± s.d.
Extended Data Fig. 2 Design and optimization of a ROSALIND reaction.
Constructing a ROSALIND reaction begins with identifying a ligand of interest, an aTF capable of binding or unbinding a DNA operator sequence as a function of the ligand and the aTF’s cognate operator sequence. Once identified, two DNA constructs are designed, one to separately express and purify the aTF and the other to generate a linear transcription template encoding the promoter, spacer, operator, fluorescent RNA aptamer and optional terminator. The first step in optimizing the sensor is performing a titration of the purified aTF at a fixed DNA template concentration to determine the amount of aTF needed to fully repress the expression of 3WJdB. Then, the target ligand is titrated (at the concentration of aTF previously determined) to test for induction. If the sensor needs improvement, components of ROSALIND can be redesigned and retested. The data shown here are for the TetR-based ROSALIND reaction described in Fig. 2f,h. The bar graphs shown are data taken at 4 h after initiating reactions with T7 RNAP and 25 nM DNA template, and the kinetics data are shown for 0 µM (green lines) and 1.25 µM TetR dimer (purple lines). Fluorescence-activation is substantially repressed at 25-fold excess or greater of TetR dimer over DNA template. The TetR-aTc dose response curve shown are the data presented in Fig. 2h, and the kinetics data are shown for 0 µM (purple lines) and 2.5 µM aTc (green lines). All data shown for n = 3 independent biological replicates as points or lines with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Shading and error bars indicate the average value of 3 independent biological replicates ± s.d.
Extended Data Fig. 3 Characterization of a ROSALIND dose response curve.
A dose response curve of a TetR-regulated ROSALIND reaction is plotted from the measured and calibrated kinetics traces, using the 4-h end point fluorescence values at different ligand concentrations. a, kinetic traces of TetR–aTc induction at different aTc concentrations, and b, the corresponding 4-h fluorescence values plotted against the µM of aTc added. Reactions are generated using 25 nM of DNA and 1.25 µM of TetR dimer. All data shown for n = 3 independent biological replicates as lines (a) or points (b) with raw fluorescence values standardized to MEF (µM FITC). Shading (a) and error bars (b) indicate the average value of 3 independent biological replicates ± s.d.
Extended Data Fig. 4 Dose response curves of ROSALIND reactions.
The dose response curves of ROSALIND reactions shown in Fig. 3 are presented. The amount of DNA templates and aTF used in each panel are configured as described in Supplementary Table 2. All data shown for n = 3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± s.d. The ligand concentrations at which the signal is distinguishable from the background were determined using two-tailed heteroscedastic Student’s t-test against the no ligand condition, and their p-value ranges are indicated with black asterisks (*** < 0.001, ** = 0.001–0.01, * = 0.01–0.05). The asterisks indicated in red have p-values in the range between 0.05–0.15 due to variability between replicates, although the average signals were clearly above background. Exact p-values along with degrees of freedom can be found in Supplementary Data 3. Data for no ligand conditions were excluded in the plots since the x-axis is on the log scale, but they can be found in Supplementary Data 3.
Extended Data Fig. 5 Orthogonality of ROSALIND reactions.
The corresponding bar graph data of the orthogonality matrix shown in Fig. 4a are presented. All data shown for n = 3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± s.d. Tet, Tetracycline; Ery, Erythromycin; 3-OH BA, 3-Hydroxybenzoic Acid; BAC, Benzalkonium chloride.
Extended Data Fig. 6 RNA kleptamers are used to invert transcription factor responses.
a, A kleptamer RNA (KB2) antisense to the dye-binding region of the broccoli aptamer can disrupt folding of 3WJdB and lead to the loss of fluorescence. Addition of a KB2-expressing template in a 4:1 ratio with the 3WJdB template inhibits signal. b, KB2 transcription can be regulated by TetR by placing the tetO site in between the T7 promoter and KB2 coding sequence. In the presence of 1.25 μM TetR dimer, the KB2 transcription is fully repressed, and the signal from 3WJdB is restored. c, Kleptamers can be used to invert the response of transcription factors when KB2 is regulated by TetR (1.25 μM dimer) and induced by aTc (2.5 μM shown). d, This scheme was used to create a ROSALIND zinc sensor with the aporepressor AdcR. When bound to Zn2+ (30 μM), AdcR (1.5 μM dimer) binds to its cognate operator sequence, adcO, placed upstream of the KB2 coding sequence, preventing KB2 expression and thereby activating fluorescence from 3WJdB. Arrows inside of the plots represent direction of regulation when indicated species are added. All data shown for n = 3 independent biological replicates as lines with raw fluorescence values standardized to MEF (µM FITC). Shading indicates the average value of 3 independent biological replicates ± s.d. 3WJdB template concentrations used are: 25 nM for a-c and 7.5 nM for d. KB2 template concentrations used are: 100 nM for a, and 150 nM for b-d.
Extended Data Fig. 7 ROSALIND sensitivity can be tuned by changing the concentration of transcription factor.
a, Increasing the concentration of CsoR desensitizes the copper sensor (dose response shift to higher concentrations), while b, decreasing the concentrations of CadC sensitizes the lead sensor (dose response shift to lower concentrations). The red arrows in the plots indicate the direction of dose response shift. All data shown for n = 3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC). Error bars indicate the average value of 3 independent biological replicates ± s.d. 3WJdB DNA template concentrations used are: 25 nM for a and 1.5 µM CadC condition in b, and 10 nM for 0.5 µM CadC condition in b.
Extended Data Fig. 8 Stability of ROSALIND.
a, The fluorescence signal generated from unregulated ROSALIND reactions are stable over weeks. The increase in fluorescence from Day 1 is likely due to a concentration increase caused by evaporation when the plate was taken out of the incubator after the first measurements. b, The shelf-stability of freeze-dried ROSALIND reactions (unregulated, TetR-regulated and aTc-induced) decay over the course of a month without proper packaging. c, Packaging of freeze-dried ROSALIND: 1) reactions are lyophilized overnight, 2) the overnight lyophilized reactions are purged with inert gas such as argon and 3) the reactions are placed into a light-protective bag with a desiccant and immediately impulse heat sealed (Supplementary Video 2). d, When this packaging method is implemented, lyophilized reactions are functional out to 2.5 months. Though we observed signal decay, the signal from rehydrated reactions after 2.5 months is clearly visible. Images are shown for one replicate with other replicate images included in Supplementary Data 1. Unregulated reactions were lyophilized with 25 nM of the 3WJdB template, and TetR-regulated reactions with additional 1.25 µM TetR dimer along with the components of IVT specified in the In vitro transcription reactions method section. Unregulated and TetR-regulated reactions were then rehydrated with laboratory-grade water, and aTc-induced reactions were rehydrated with 10 µM of aTc. All data shown for n = 3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and center values representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± s.d. The original, uncropped images shown in c and d can be found in Supplementary Data 1.
Extended Data Fig. 9 Handheld fluorescence illuminator design.
a, A low-cost handheld fluorescence illuminator enables point-of-use functionality for ROSALIND. 3D printed components include a, b, front case, c, back case and d, battery cover. A printed circuit board (e, front view facing back case, f, rear view facing front case) mounts in the front case and connects LEDs, resistors, a trimmer potentiometer, a power switch and a battery holder. Full CAD files, 3D print files (.STL), PCB design files and assembly instructions are provided in Supplementary Data 4. The estimated cost of building one device is $8.69 USD.
Extended Data Fig. 10 ROSALIND reactions can be freeze-dried and rehydrated with an environmental surface water source.
a, The ROSALIND reactions shown in Fig. 6 were characterized for response to CuSO4 as in Supplementary Fig. 6, but using water from Lake Michigan (collected in Evanston, IL). We observed lower signals from these spiked environmental samples than with the laboratory-grade water samples due to uncharacterized matrix effects. b, To test ROSALIND functionality on real-world environmental samples, surface freshwater samples were obtained from two streams known to contain elevated levels of copper: (1) the Yerba Loca Creek, upstream from Santiago, in the Maipo watershed of Central Chile, and (2) the Toro River, upstream from La Serena, in the Elqui watershed of Northern Chile. c, d, Copper concentrations of the samples from the Yerba Loca Creek (c) and the Toro River (d) were determined using Flame Atomic Absorption Spectroscopy (FAAS) and found to be in the range of 6.9–8.5 ppm. FAAS was calibrated with a Cu(NO3)2 standard solution as shown in e. Serial dilution was performed on each sample to create three separate diluted samples that are in the operating ppm range of FAAS, and their ppm values were measured and reported in Table f as the average value of 3 independent replicates ± s.d. Using these measurements, linear regression on the averages of each dilution was performed to calculate the ppm value of the undiluted sample. g, h, In order to generate a series of tests that cover our detectable copper range, we serially diluted each sample with laboratory-grade, metal-free water (undiluted, 1:2, 1:4, 1:8, 1:16 and 1:32 dilutions) and measured their concentrations by FAAS before using this series to rehydrate freeze-dried copper sensors. The copper concentrations indicated were taken from c, d, or calculated from the extrapolated ppm value of the undiluted sample in Table f. When rehydrated with the 1:8 dilution, we observed clear visible signals, corresponding to copper concentrations of 0.88 ppm and 1.05 ppm from the Yerba Loca Creek and Toro River samples, respectively. Four-hour end-point data are shown in a, g, h for n = 3 independent biological replicates as points with raw fluorescence values standardized to MEF (µM FITC), and bars representing averages of the replicates. Error bars indicate the average value of 3 independent biological replicates ± s.d. Geographical data in b © OpenStreetMap contributors46.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Tables 1–5 and References.
41587_2020_571_MOESM3_ESM.mov
Supplementary Video 1 Illumination of an unregulated ROSALIND reaction. The video shows two ROSALIND reactions with and without 250 nM 3WJdB DNA template run using the NEB HiScribe T7 Quick High Yield RNA Synthesis Kit. The reactions were incubated at 37 °C and monitored in real time using the 3D-printed, handheld illuminator. The speed of the video has been sped up 90× to show the 90-min-long incubation in a 1-min period.
41587_2020_571_MOESM4_ESM.mov
Supplementary Video 2 Freeze-drying and packaging of ROSALIND reactions. The video shows the method used to freeze-dry and package ROSALIND reactions. ROSALIND reactions are prepared on a sterilized aluminum block and sealed with a cap with three holes. A lyophilizing jar is prepared by padding its inside with tissues and wrapping its outside with aluminum foil to protect from light. Reactions are then flash-frozen on the aluminum block using liquid nitrogen, wrapped with tissues and aluminum foil, placed inside the lyophilizing jar and lyophilized overnight. Lyophilized reactions are then removed from the jar, put inside a vacuum-sealable bag with several desiccants, purged with argon gas and sealed immediately. The vacuum-sealed reactions are placed inside a light-protective bag and stored in a cool, shaded area until usage.
Supplementary Data 1
Raw illuminator and freeze-dried packaging method images included in figures.
Supplementary Data 2
Template Excel worksheet for setting up a ROSALIND reaction.
Supplementary Data 3
Calibrated plate reader data for all figures.
Supplementary Data 4
Instructions for 3D printing and assembling a handheld illuminator.
Supplementary Data 5
DNA and protein sequences used in the present study.
Source data
Source Data Fig. 2
Fig. 2b,d,f,h Calibrated plate reader data.
Source Data Fig. 3
Fig. 3 Calibrated plate reader data.
Source Data Fig. 4
Fig. 4a,c Calibrated plate reader data.
Source Data Fig. 5
Fig. 5b Calibrated plate reader data and statistical source data.
Source Data Fig. 6
Fig. 6c–f Calibrated plate reader data.
Source Data Extended Data Fig. 1
Calibrated plate reader data.
Source Data Extended Data Fig. 2
Calibrated plate reader data.
Source Data Extended Data Fig. 3
Calibrated plate reader data.
Source Data Extended Data Fig. 4
Calibrated plate reader data and statistical source data.
Source Data Extended Data Fig. 5
Calibrated plate reader data.
Source Data Extended Data Fig. 6
Calibrated plate reader data.
Source Data Extended Data Fig. 7
Calibrated plate reader data.
Source Data Extended Data Fig. 8
Extended Data Fig. 8a,b,d Calibrated plate reader data
Source Data Extended Data Fig. 10
Extended Data Fig. 10a,g,h Calibrated plate reader data.
Rights and permissions
About this article
Cite this article
Jung, J.K., Alam, K.K., Verosloff, M.S. et al. Cell-free biosensors for rapid detection of water contaminants. Nat Biotechnol 38, 1451–1459 (2020). https://doi.org/10.1038/s41587-020-0571-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0571-7
This article is cited by
-
Snowprint: a predictive tool for genetic biosensor discovery
Communications Biology (2024)
-
Use of antioxidants to extend the storage of lyophilized cell-free synthesis system
Biotechnology and Bioprocess Engineering (2024)
-
Automated design of protein-binding riboswitches for sensing human biomarkers in a cell-free expression system
Nature Communications (2023)
-
The accuracy and usability of point-of-use fluoride biosensors in rural Kenya
npj Clean Water (2023)
-
Voltammetric investigation of the complexing effect of Capparis spinosa on heavy metals: Application in the treatment of water
Ionics (2023)