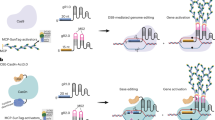
Abstract
Short insertions and deletions can be produced in plant genomes using CRISPR–Cas editors, but reliable production of larger deletions in specific target sites has proven difficult to achieve. We report the development of a series of APOBEC–Cas9 fusion-induced deletion systems (AFIDs) that combine Cas9 with human APOBEC3A (A3A), uracil DNA-glucosidase and apurinic or apyrimidinic site lyase. In rice and wheat, AFID-3 generated deletions from 5′-deaminated C bases to the Cas9-cleavage site. Approximately one-third of deletions produced using AFID-3 in rice and wheat protoplasts (30.2%) and regenerated plants (34.8%) were predictable. We show that eAFID-3, in which the A3A in AFID-3 is replaced with truncated APOBEC3B (A3Bctd), produced more uniform deletions from the preferred TC motif to the double-strand break. AFIDs could be applied to study regulatory regions and protein domains to improve crop plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
NGS data have been deposited in the NCBI Sequence Read Archive database (accession no. PRJNA630559). Two plasmids encoding AFID-3 and eAFID-3 in the present study will be available through Addgene. All data supporting the findings of the present study are available in the article and its supplementary figures and tables, or from the corresponding author on request. For sequence data, OsAAT (LOC_Os01g55540), OsACC (LOC_Os05g22940), OsCDC48 (LOC_Os03g05730), OsEV (LOC_Os02g11010), OsNRT1.1B (LOC_Os10g40600), OsSPL14/OsIPA1 (LOC_Os08g39890) and OsSWEET14 (LOC_Os11g31190) are from Rice Genome Annotation Project (http://rice.plantbiology.msu.edu); TaMYB10 (AB191458.1, AB191459.1, AB191460.1) and TaVRN1 (AY747603.1, AY747604.1, AY747605.1) are from the NCBI (https://www.ncbi.nlm.nih.gov); TaF3H (TraesCS2A02G493500, TraesCS2B02G521500, TraesCS2D02G493400), TaGASR6 (TraesCS1A02G270100, TraesCS1D02G270100), TaPDS (TraesCS4A02G004900, TraesCS4B02G300100, TraesCS4D02G299000) and TaPMK (TraesCS5A02G449000, TraesCS5B02G453800, TraesCS5D02G455500) are from the International Wheat Genome Sequencing Consortium (https://urgi.versailles.inra.fr); and tae-Pri-miR160, tae-Pri-miR319, tae-Pri-miR396 and tae-Pri-miR444a are from miRBase (http://www.mirbase.org).
References
Knott, G. J. & Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Chen, K., Wang, Y., Zhang, R., Zhang, H. & Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017).
Stinson, B. M., Moreno, A. T., Walter, J. C. & Loparo, J. J. A mechanism to minimize errors during non-homologous end joining. Mol. Cell 77, 1080–1091 (2020).
Taheri-Ghahfarokhi, A. et al. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 46, 8417–8434 (2018).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Burgess, D. G., Xu, J. & Freeling, M. Advances in understanding cis regulation of the plant gene with an emphasis on comparative genomics. Curr. Opin. Plant Biol. 27, 141–147 (2015).
Li, C. et al. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotech. J. 18, 313–315 (2020).
Li, M. et al. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7, 377 (2016).
Zhang, X., Wang, L., Liu, M. & Li, D. CRISPR/Cas9 system: a powerful technology for in vivo and ex vivo gene therapy. Sci. China Life Sci. 60, 468–475 (2017).
Zhou, J. et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 8, 1598 (2017).
Bolukbasi, M. F. et al. Orthogonal Cas9–Cas9 chimeras provide a versatile platform for genome editing. Nat. Commun. 9, 4856 (2018).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Shou, J., Li, J., Liu, Y. & Wu, Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol. Cell 71, 498–509 (2018).
Wolfs, J. M. et al. Biasing genome-editing events toward precise length deletions with an RNA-guided TevCas9 dual nuclease. Proc. Natl Acad. Sci. USA 113, 14988–14993 (2016).
Čermák, T. et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29, 1196–1217 (2017).
Zhang, Q., Yin, K., Liu, G., Li, S. & Qiu, J. L. Fusing T5 exonuclease with Cas9 and Cas12a increases the frequency and size of deletion at target sites. Sci. China Life Sci. https://doi.org/10.1007/s11427-020-1671-6 (2020).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Lin, Q. et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Lei, L. et al. APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks. Nat. Struct. Mol. Biol. 25, 45–52 (2018).
Que, Q. et al. Plant DNA repair pathways and their applications in genome engineering. Methods Protoc. 1917, 3–24 (2019).
Zong, Y. et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953 (2018).
Gisler, S. et al. Multiplexed Cas9 targeting reveals genomic location effects and gRNA-based staggered breaks influencing mutation efficiency. Nat. Commun. 10, 1598 (2019).
Zuo, Z. & Liu, J. Cas9-catalyzed DNA cleavage generates staggered ends: evidence from molecular dynamics simulations. Sci. Rep. 5, 37584 (2016).
Li, T., Liu, B., Spalding, M. H., Weeks, D. P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Oliva, R. et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350 (2019).
Xu, Z. et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446 (2019).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Gehrke, J. M. et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977–982 (2018).
St. Martin, A. et al. A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res. 46, e84 (2018).
Siriwardena, S. U., Guruge, T. A. & Bhagwat, A. S. Characterization of the catalytic domain of human APOBEC3B and the critical structural role for a conserved methionine. J. Mol. Biol. 427, 3042–3055 (2015).
Omidbakhshfard, M., Proost, S., Fujikura, U. & Mueller-Roeber, B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010 (2015).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Allen, F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37, 64–72 (2018).
Leenay, R. T. et al. Large dataset enables prediction of repair after CRISPR-Cas9 editing in primary T cells. Nat. Biotechnol. 37, 1034–1037 (2019).
Turonyi, B. W. et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079 (2019).
He, W. et al. De novo identification of essential protein domains from CRISPR-Cas9 tiling-sgRNA knockout screens. Nat. Commun. 46, 505–529 (2019).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Shan, Q. et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Zhang, Y., Zhang, Q. & Chen, Q. J. Agrobacterium-mediated delivery of CRISPR/Cas reagents for genome editing in plants enters an era of ternary vector systems. Sci. China Life Sci. https://doi.org/10.1007/s11427-020-1685-9 (2020).
Acknowledgements
We thank Professor G. Chen (Shanghai Jiao Tong University) for providing the PH strain containing the TAL effector AvaXa7 for evaluation of bacterial blight resistance. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, grant no. XDA24020100), the National Transgenic Science and Technology Program (grant no. 2016ZX08010002), the National Natural Science Foundation of China (grant no. 31788103, 31971370), the National Key Research and Development Program of China (grant no. 2016YFD0101804), the Chinese Academy of Sciences (grant no. QYZDY-SSW-SMC030) and the Postdoctoral Innovative Talent Support Program of China (grant no. BX20190365).
Author information
Authors and Affiliations
Contributions
S.W., H.Z. and C.G. designed the project. S.W., Y.Z., Q.L., H.Z., Z.C., K.C. and D.Z. performed the experiments. C.G. supervised the project. S.W., Y.Z., Q.L., H.Z, J.-L.Q. and C.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have submitted a patent application based on the results reported in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Tables 1 and 2 and Sequences 1 and 2.
Rights and permissions
About this article
Cite this article
Wang, S., Zong, Y., Lin, Q. et al. Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC–Cas9. Nat Biotechnol 38, 1460–1465 (2020). https://doi.org/10.1038/s41587-020-0566-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0566-4
This article is cited by
-
Application of CRISPR/Cas system in cereal improvement for biotic and abiotic stress tolerance
Planta (2022)
-
Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit
Nature Plants (2021)
-
Precise plant genome editing using base editors and prime editors
Nature Plants (2021)
-
Genome editing for crop improvement: A perspective from India
In Vitro Cellular & Developmental Biology - Plant (2021)
-
Generating novel plant genetic variation via genome editing to escape the breeding lottery
In Vitro Cellular & Developmental Biology - Plant (2021)