Abstract
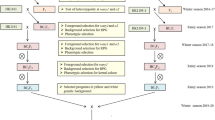
We created waxy corn hybrids by CRISPR–Cas9 editing of a waxy allele in 12 elite inbred maize lines, a process that was more than a year faster than conventional trait introgression using backcrossing and marker-assisted selection. Field trials at 25 locations showed that CRISPR-waxy hybrids were agronomically superior to introgressed hybrids, producing on average 5.5 bushels per acre higher yield.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data supporting the findings of this study are available in the article and its Supplementary Information files. Sequencing data are available in the SRA database in BioProject PRJNA597019. Plasmid accession numbers are MN294713 (Plasmid 1), MN294714 (Plasmid 2), MN294715 (Plasmid 3), MN294716 (Plasmid 4), MN294717 (Plasmid 5), MN294718 (Plasmid 6) and MN294719 (Plasmid2_Waxy-CR2). Upon request, maize lines can be provided under an applicable material transfer agreement to academic investigators for noncommercial research.
References
Shure, M., Wessler, S. & Fedoroff, N. Cell 35, 225–233 (1983).
Nelson, O. & Pan, D. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 475–496 (1995).
Schwatz, D. & Whistler, R. L. in Food Science and Technology (eds BeMiller, J. & Whistler, R.) 1–10 (Academic, 2009).
Fan, L. et al. PLoS ONE 4, e7612 (2009).
Dong, L. et al. Plant Biotechnol. J. 17, 1853–1855 (2019).
Svitashev, S. et al. Plant Physiol. 169, 931–945 (2015).
Lowe, K. et al. In Vitro Cell Dev. Biol. Plant 54, 240–252 (2018).
Chilcoat, D., Liu, Z. B. & Sander, J. Prog. Mol. Biol. Transl. Sci. 149, 27–46 (2017).
Shi, J. et al. Plant Biotechnol. J. 15, 207–216 (2017).
Zastrow-Hayes, G. et al. Plant Genome 8, 1–15 (2015).
Young, J. et al. Sci. Rep. 9, 6729 (2019).
Tang, X. et al. Genome Biol. 19, 84 (2018).
Hahn, F. & Nekrasov, V. Plant Cell Rep. 38, 437–441 (2019).
Li, J. et al. Plant Biotechnol. J. 17, 858–868 (2019).
Gao, H. et al. Plant J. 61, 176–187 (2010).
McCleary, B. V., Solah, V. & Gibson, T. S. J. Cereal Sci. 20, 51–58 (1994).
Gilmour, A. R., Thompson, R. & Cullis, B. R. Biometrics 51, 1440–1450 (1995).
Cullis, B. R. et al. Biometrics 54, 1–18 (1998).
Gilmour, A. R. et al. ASReml User Guide 3.0. (VSN International, 2009).
Acknowledgements
We thank D. O’Neil, A. Carzoli, G. Zastrow-Hayes and their teams for nucleic acid analysis and sequencing; L. Church, K. Simcox and their team for greenhouse plant care and nusery seed production; and S. Basu, S. Betts and D. Bubeck for providing project support. We are grateful to J. Shi, S. Betts, G. May and M. Fedorova for critical review of this manuscript.
Author information
Authors and Affiliations
Contributions
R.B.M., H.G., A.M.C., N.D.C. and T.W.G. designed edits. M.J.G. selected inbred lines. H.G., B.L., M.Y., M.S., J.F., K.S., D.P., L.F., S.J., G.St.C., M.R., N.S.-D., C.P., L.W., J.K.Y., M.B. and J.H. conducted the transformation and molecular analysis experiments. M.J.G., H.R.L. and B.D. conducted field trials and analyzed data. H.G., M.J.G. and N.D.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
This work was funded by Corteva Agriscience, a for-profit agricultural technology company, as part of its research and development program. All authors were employees of Corteva Agriscience at the time of their contributions to this work. Patents have been filed related to this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Materials
Supplementary Figs. 1–8 and Tables 1–7.
Rights and permissions
About this article
Cite this article
Gao, H., Gadlage, M.J., Lafitte, H.R. et al. Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat Biotechnol 38, 579–581 (2020). https://doi.org/10.1038/s41587-020-0444-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0444-0
This article is cited by
-
Application of nanotechnology and proteomic tools in crop development towards sustainable agriculture
Journal of Crop Science and Biotechnology (2024)
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Targeted genome editing for cotton improvement: prospects and challenges
The Nucleus (2024)
-
Genome editing in cotton: challenges and opportunities
Journal of Cotton Research (2023)
-
Leaf transformation for efficient random integration and targeted genome modification in maize and sorghum
Nature Plants (2023)