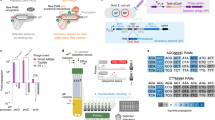
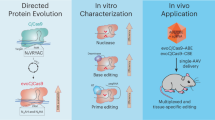
Abstract
The targeting scope of Streptococcus pyogenes Cas9 (SpCas9) and its engineered variants is largely restricted to protospacer-adjacent motif (PAM) sequences containing G bases. Here we report the evolution of three new SpCas9 variants that collectively recognize NRNH PAMs (where R is A or G and H is A, C or T) using phage-assisted non-continuous evolution, three new phage-assisted continuous evolution strategies for DNA binding and a secondary selection for DNA cleavage. The targeting capabilities of these evolved variants and SpCas9-NG were characterized in HEK293T cells using a library of 11,776 genomically integrated protospacer–sgRNA pairs containing all possible NNNN PAMs. The evolved variants mediated indel formation and base editing in human cells and enabled A•T-to-G•C base editing of a sickle cell anemia mutation using a previously inaccessible CACC PAM. These new evolved SpCas9 variants, together with previously reported variants, in principle enable targeting of most NR PAM sequences and substantially reduce the fraction of genomic sites that are inaccessible by Cas9-based methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
HTS data were deposited in the Sequence Read Archive database (PRJNA596996). Plasmids encoding SpCas9-NRRH, SpCas9-NRTH and SpCas9-NRCH nucleases, BE4 and ABE are available through Addgene. A subset of the selection plasmids used in this study are available through Addgene. Other materials are available from the corresponding authors upon reasonable request.
Code availability
Custom scripts used to analyze the human cell target site library are available at https://github.com/maxwshen-PAMvar-processing.
References
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cebrian-Serrano, A. & Davies, B. CRISPR–Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm. Genome 28, 247–261 (2017).
Kleinstiver, B. P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR–Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Nishimasu, H. et al. Engineered CRISPR–Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Iyer, S. et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565 (2019).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–778 (2018).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Xin, H., Wan, T. & Ping, Y. Off-targeting of base editors: BE3 but not ABE induces substantial off-target single nucleotide variants. Signal Transduct. Target. Ther. 4, 9 (2019).
Lee, H. K. et al. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat. Commun. 9, 4804 (2018).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Dickinson, B. C., Leconte, A. M., Allen, B., Esvelt, K. M. & Liu, D. R. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc. Natl Acad. Sci. USA 110, 9007–9012 (2013).
Carlson, J. C., Badran, A. H., Guggiana-nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10, 216–222 (2014).
Pu, J., Disare, M. & Dickinson, B. C. Evolution of C-terminal modification tolerance in full-length and split T7 RNA polymerase biosensors. Chembiochem 20, 1547–1553 (2019).
Pu, J., Kentala, K. & Dickinson, B. C. Multidimensional control of Cas9 by evolved RNA polymerase-based biosensors. ACS Chem. Biol. 13, 431–437 (2018).
Pu, J., Zinkus-Boltz, J. & Dickinson, B. C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 13, 432–438 (2017).
Packer, M. S., Rees, H. A. & Liu, D. R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 8, 956 (2017).
Dickinson, B. C., Packer, M. S., Badran, A. H. & Liu, D. R. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat. Commun. 5, 5352 (2014).
Hubbard, B. P. et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Methods 12, 939–942 (2015).
Badran, A. H. et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533, 58–63 (2016).
Wang, T., Badran, A. H., Huang, T. P. & Liu, D. R. Continuous directed evolution of proteins with improved soluble expression. Nat. Chem. Biol. 14, 972–980 (2018).
Bryson, D. I. et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 13, 1253–1260 (2017).
Suzuki, T. et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 13, 1261–1266 (2017).
Roth, T. B., Woolston, B. M., Stephanopoulos, G. & Liu, D. R. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth. Biol. 8, 796–806 (2019).
Karvelis, T. et al. Rapid characterization of CRISPR–Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253 (2015).
Shah, N. H., Dann, G. P., Vila-Perelló, M., Liu, Z. & Muir, T. W. Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 (2012).
Brödel, A. K., Jaramillo, A. & Isalan, M. Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat. Commun. 7, 13858 (2016).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Jiang, F. & Doudna, J. A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529 (2017).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Guo, M. et al. Structural insights into a high fidelity variant of SpCas9. Cell Res. 29, 183–192 (2019).
Kim, S., Bae, T., Hwang, J. & Kim, J. S. Rescue of high-specificity Cas9 variants using sgRNAs with matched 5′ nucleotides. Genome Biol. 18, 218 (2017).
Lee, J. K. et al. Directed evolution of CRISPR–Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Zhang, D. et al. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 18, 191 (2017).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Rees, D. C., Williams, T. N. & Gladwin, M. T. Sickle-cell disease. Lancet 376, 2018–2031 (2010).
Blackwell, R. Q. et al. Hemoglobin G Makassar: β6 Glu→Ala. Biochim. Biophys. Acta. 214, 396–401 (1970).
Viprakasit, V., Wiriyasateinkul, A., Sattayasevana, B., Miles, K. L. & Laosombat, V. Hb G-Makassar [β6(A3)Glu→Ala; codon 6 (GAG→GCG)]: molecular characterization, clinical, and hematological effects. Hemoglobin 26, 245–253 (2002).
Sangkitporn, S. et al. Hb G Makassar (β6: Glu-Ala) in a Thai family. J. Med. Assoc. Thai 85, 577–582 (2002).
Hirano, S., Nishimasu, H., Ishitani, R. & Nureki, O. Structural basis for the altered PAM specificities of engineered CRISPR–Cas9. Mol. Cell 61, 886–894 (2016).
Anders, C., Bargsten, K. & Jinek, M. Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol. Cell 61, 895–902 (2016).
Chatterjee, P., Jakimo, N. & Jacobson, J. M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 4, eaau0766 (2018).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Esvelt, K. M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726 (2019).
Hirano, H. et al. Structure and engineering of Francisella novicida Cas9. Cell 164, 950–961 (2016).
Harrington, L. B. et al. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 8, 1424 (2017).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771 (2015).
Hou, Z. et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl Acad. Sci. USA 110, 15644–15649 (2013).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Badran, A. H. & Liu, D. R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 6, 8425 (2015).
Peng, X., Nguyen, A. & Ghosh, D. Quantification of M13 and T7 bacteriophages by TaqMan and SYBR green qPCR. J. Virol. Methods 242, 100–107 (2018).
Gaudelli, N. M. et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Müller, K. M. et al. Nucleotide exchange and excision technology (NExT) DNA shuffling: a robust method for DNA fragmentation and directed evolution. Nucleic Acids Res. 33, e117 (2005).
Dower, W. J., Miller, J. F. & Ragsdale, C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145 (1988).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Arbab, M., Srinivasan, S., Hashimoto, T., Geijsen, N. & Sherwood, R. I. Cloning-free CRISPR. Stem Cell Rep. 5, 908–917 (2015).
Urasaki, A., Morvan, G. & Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006).
Acknowledgements
We thank T.R. Blum and A. Raguram for helpful discussions. This work was supported by National Institutes of Health grants R01 EB027793, U01 AI142756, RM1 HG009490 and R35 GM118062, the Howard Hughes Medical Institute, the Bill and Melinda Gates Foundation and the St. Jude Collaborative Research Consortium. S.M.M. and M.S. were supported by a National Science Foundation Graduate Research Fellowship. T.W. was supported by a Ruth L. Kirchstein National Research Service Awards Postdoctoral Fellowship (F32GM119228). M.A. was supported by an NWO Rubicon Fellowship. G.A.N was supported by a Helen Hay Whitney Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
S.M.M., T.W. and D.R.L. designed the research. S.M.M., T.W. and P.B.R. performed evolution. S.M.M., T.W., P.B.R. and T.P.H. characterized variants in bacteria. S.M.M., T.W., P.B.R., M.A. and Z.M. conducted human cell experiments. M.A. and M.S. designed and constructed the integrated target site human cell library. M.S. wrote custom software. S.M.M., T.W., M.A. and M.S. analyzed library data. S.M.M. and T.W. performed GUIDE-seq experiments. G.N. and H.A.R. designed and performed the sickle cell anemia site experiments. S.M.M., T.W., P.B.R. and T.P.H. designed and performed the HbS editing experiments. S.M.M., T.W. and D.R.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare competing financial interests: S.M.M., T.W. and D.R.L. have filed patent applications on this work. D.R.L. is a consultant and co-founder of Editas Medicine, Pairwise Plants, Beam Therapeutics and Prime Medicine, companies that use genome-editing technologies.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1, 3 and 6–9, and Notes 1–4
Supplementary Table 2
Excel table of PACE mutations.
Supplementary Table 4
Excel table of GUIDE-seq-identified off-target sites.
Supplementary Table 5
Excel table of PAMvar library data on all sites.
Rights and permissions
About this article
Cite this article
Miller, S.M., Wang, T., Randolph, P.B. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol 38, 471–481 (2020). https://doi.org/10.1038/s41587-020-0412-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0412-8
This article is cited by
-
Ultrahigh-throughput screening-assisted in vivo directed evolution for enzyme engineering
Biotechnology for Biofuels and Bioproducts (2024)
-
Phage-assisted evolution of compact Cas9 variants targeting a simple NNG PAM
Nature Chemical Biology (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Nature Communications (2024)
-
Deep learning models to predict the editing efficiencies and outcomes of diverse base editors
Nature Biotechnology (2024)