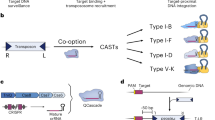
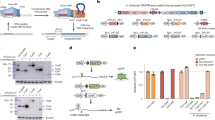
Abstract
Existing technologies for site-specific integration of kilobase-sized DNA sequences in bacteria are limited by low efficiency, a reliance on recombination, the need for multiple vectors, and challenges in multiplexing. To address these shortcomings, we introduce a substantially improved version of our previously reported Tn7-like transposon from Vibrio cholerae, which uses a Type I-F CRISPR–Cas system for programmable, RNA-guided transposition. The optimized insertion of transposable elements by guide RNA–assisted targeting (INTEGRATE) system achieves highly accurate and marker-free DNA integration of up to 10 kilobases at ~100% efficiency in bacteria. Using multi-spacer CRISPR arrays, we achieved simultaneous multiplexed insertions in three genomic loci and facile, multi-loci deletions by combining orthogonal integrases and recombinases. Finally, we demonstrated robust function in biomedically and industrially relevant bacteria and achieved target- and species-specific integration in a complex bacterial community. This work establishes INTEGRATE as a versatile tool for multiplexed, kilobase-scale genome engineering.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
NGS data are available in the NCBI Sequence Read Archive (BioProject accession code PRJNA668381). Published genomes used for analyses were obtained from the NCBI (accessions codes CP001509.3, U00096.3, CP009273.1 and AE015451.2). Datasets generated and analyzed in the current study, as well as custom scripts used for the described data analyses, are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
Custom Python scripts used for the described NGS data analyses are available online via GitHub (https://github.com/sternberglab/Vo_etal_2020). The INTEGRATE guide RNA design tool and associated documentation are available online via GitHub (https://github.com/sternberglab/INTEGRATE-guide-RNA-tool).
References
Dunbar, C. E. et al. Gene therapy comes of age. Science 359, eaan4672 (2018).
Gelvin, S. B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 51, 195–217 (2017).
Davy, A. M., Kildegaard, H. F. & Andersen, M. R. Cell factory engineering. Cell Syst. 4, 262–275 (2017).
Brophy, J. A. N. et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol. 3, 1043–1053 (2018).
Miyazaki, R. & van der Meer, J. R. A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl. Environ. Microbiol. 79, 4440–4447 (2013).
Martínez-García, E. & de Lorenzo, V. Transposon-based and plasmid-based genetic tools for editing genomes of gram-negative bacteria. Methods Mol. Biol. 813, 267–283 (2012).
van Opijnen, T., Bodi, K. L. & Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009).
Wang, H. H. et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012).
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G. & Court, D. L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009).
Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Cotta-de-Almeida, V., Schonhoff, S., Shibata, T., Leiter, A. & Snapper, S. B. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 13, 2190–2194 (2003).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Wang, K. et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016).
Sukhija, K. et al. Developing an extended genomic engineering approach based on recombineering to knock-in heterologous genes to Escherichia coli genome. Mol. Biotechnol. 51, 109–118 (2012).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Vento, J. M., Crook, N. & Beisel, C. L. Barriers to genome editing with CRISPR in bacteria. J. Ind. Microbiol. Biotechnol. 46, 1327–1341 (2019).
Jiang, Y. et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 8, 15179 (2017).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Wannier, T. M. et al. Improved bacterial recombineering by parallelized protein discovery. Proc. Natl Acad. Sci. USA 117, 13689–13698 (2020).
Corts, A. D., Thomason, L. C., Gill, R. T. & Gralnick, J. A. A new recombineering system for precise genome-editing in Shewanella oneidensis strain MR-1 using single-stranded oligonucleotides. Sci. Rep. 9, 39 (2019).
Peters, J. M. et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat. Microbiol. 4, 244–250 (2019).
St-Pierre, F. et al. One-step cloning and chromosomal integration of DNA. ACS Synth. Biol. 2, 537–541 (2013).
Tellier, M., Bouuaert, C. C. & Chalmers, R. Mariner and the ITm superfamily of transposons. Microbiol. Spectr. 3, MDNA3–0033–2014 (2015).
van Opijnen, T. & Camilli, A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013).
Haniford, D. B. & Ellis, M. J. Transposons Tn10 and Tn5. Microbiol. Spectr. 3, MDNA3–0002–2014 (2015).
Goodall, E. C. A. et al. The essential genome of Escherichia coli K-12. Mbio 9, e02096-17 (2018).
Chen, S. P. & Wang, H. H. An engineered Cas-transposon system for programmable and site-directed DNA transpositions. CRISPR J. 2, 376–394 (2019).
Bhatt, S. & Chalmers, R. Targeted DNA transposition in vitro using a dCas9-transposase fusion protein. Nucleic Acids Res. 6, 7–10 (2019).
Enyeart, P. J., Mohr, G., Ellington, A. D. & Lambowitz, A. M. Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob. DNA 5, 2 (2014).
Esvelt, K. M. & Wang, H. H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641 (2013).
Perutka, J., Wang, W., Goerlitz, D. & Lambowitz, A. M. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 336, 421–439 (2004).
Karberg, M. et al. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19, 1162–1167 (2001).
Klompe, S. E., Vo, P. L. H., Halpin-Healy, T. S. & Sternberg, S. H. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019).
Peters, J. E., Makarova, K. S., Shmakov, S. & Koonin, E. V. Recruitment of CRISPR–Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017).
Halpin-Healy, T. S., Klompe, S. E., Sternberg, S. H. & Fernández, I. S. Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 (2020).
Faure, G. et al. CRISPR–Cas in mobile genetic elements: counter-defence and beyond. Nat. Rev. Microbiol. 17, 513–525 (2019).
Peters, J. E. Targeted transposition with Tn7 elements: safe sites, mobile plasmids, CRISPR/Cas and beyond. Mol. Microbiol. 112, 1635–1644 (2019).
Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019).
Chavez, M. & Qi, L. S. Site-programmable transposition: shifting the paradigm for CRISPR–Cas systems. Mol. Cell. 75, 206–208 (2019).
Hou, Z. & Zhang, Y. Inserting DNA with CRISPR. Science 365, 25–26 (2019).
Ronda, C., Chen, S. P., Cabral, V., Yaung, S. J. & Wang, H. H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat Meth 16, 167–170 (2019).
Stellwagen, A. E. & Craig, N. L. Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16, 6823–6834 (1997).
Greene, E. C. & Mizuuchi, K. Target immunity during Mu DNA transposition. Transpososome assembly and DNA looping enhance MuA-mediated disassembly of the MuB target complex. Mol. Cell 10, 1367–1378 (2002).
Hagemann, A. T. & Craig, N. L. Tn7 transposition creates a hotspot for homologous recombination at the transposon donor site. Genetics 133, 9–16 (1993).
Lin, M. T. et al. In Methods in Enzymology Isotope Labeling of Biomolecules—Labeling Methods Vol. 565 (Ed. Kelman, Z.) 45–66 (Academic Press, 2015).
Hickman, A. B. & Dyda, F. DNA transposition at work. Chem. Rev. 116, 12758–12784 (2016).
Abbas, A. F., Al-Saadi, A. G. M. & Alkhudhairy, M. K. Biofilm formation and virulence determinants of Klebsiella oxytoca clinical isolates from patients with colorectal cancer. J. Gastrointest. Cancer 51, 855–860 (2019).
Kim, D.-K. et al. Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2,3-butanediol production. J. Biosci. Bioeng. 116, 186–192 (2013).
Loeschcke, A. & Thies, S. Pseudomonas putida—a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214 (2015).
Nikel, P. I. & de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab. Eng. 50, 142–155 (2018).
Sun, J. et al. Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb. Cell Fact. 17, 41 (2018).
Wirth, N. T., Kozaeva, E. & Nikel, P. I. Accelerated genome engineering of Pseudomonas putida by I-SceI-mediated recombination and CRISPR–Cas9 counterselection. Microb. Biotechnol. 13, 233–249 (2020).
Tsai, S. Q. & Joung, J. K. Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 (2016).
Zhang, Y. et al. Multicopy chromosomal integration using CRISPR-associated transposases. ACS Synth. Biol. 9, 1998–2008 (2020).
Yu, B. J. & Kim, C. Minimization of the Escherichia coli genome using the Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 20, 1018–1023 (2008).
Adiego-Pérez, B. et al. Multiplex genome editing of microorganisms using CRISPR–Cas. FEMS Microbiol. Lett. 366, fnz086 (2019).
Horlbeck, M. A. et al. Mapping the genetic landscape of human cells. Cell 174, 953–967(2018).
Bassalo, M. C. et al. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol. 5, 561–568 (2016).
Rubin, B. E. et al. Targeted genome editing of bacteria within microbial communities. Preprint at bioRxiv https://doi.org/10.1101/2020.07.17.209189 (2020).
Valderrama, J. A., Kulkarni, S. S., Nizet, V. & Bier, E. A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat. Commun. 10, 5726–5728 (2019).
Duque, E. et al. Identification and elucidation of in vivo function of two alanine racemases from Pseudomonas putida KT2440. Environ. Microbiol. Rep. 9, 581–588 (2017).
Aparicio, T., de Lorenzo, V. & Martínez-García, E. CRISPR/Cas9-enhanced ssDNA recombineering for Pseudomonas putida. Microb. Biotechnol. 12, 1076–1089 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Rice, P. A., Craig, N. L. & Dyda, F. Comment on ‘RNA-guided DNA insertion with CRISPR-associated transposases’. Science 368, eabb2022 (2020).
Strecker, J., Ladha, A., Makarova, K. S., Koonin, E. V. & Zhang, F. Response to comment on ‘RNA-guided DNA insertion with CRISPR-associated transposases’. Science 368, eabb2920 (2020).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Acknowledgements
We thank N. Jaber for laboratory support, J. Bondy-Denomy for discussions, L.F. Landweber for qPCR instrument access, J. Mohabir for assistance with NGS read alignment, the JP Sulzberger Columbia Genome Center for NGS support and M.L. Smith, I. Oussenko and the Genomics Technology Laboratory at the Icahn School of Medicine at Mount Sinai for SMRT sequencing. H.H.W. acknowledges funding support for this work from the National Science Foundation (MCB-1453219), the National Institutes of Health (1U01GM110714 and 1R01AI132403), the Office of Naval Research (N00014-17-1-2353) and the Burroughs Wellcome Fund (PATH1016691). C.R. is supported by a Junior Fellows Scholarship from the Simons Society of Fellows. S.H.S. acknowledges a generous startup package from the Columbia University Irving Medical Center Dean’s Office and the Vagelos Precision Medicine Fund.
Author information
Authors and Affiliations
Contributions
P.L.H.V. and S.H.S. conceived of and designed the project, with input from C.R. and H.H.W. P.L.H.V. performed experiments and analyzed data for most E. coli experiments. C.R. performed experiments and analyzed data in K. oxytoca, P. putida and complex bacterial communities, with input from H.H.W. S.E.K. performed target immunity, ShoINT and random fragmentation NGS experiments. E.E.C. helped with cloning and transposition experiments. C.A. assisted with computational analyses of NGS data and the guide RNA design algorithm. P.L.H.V., S.H.S. and all other authors discussed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.L.H.V., S.E.K. and S.H.S. are inventors on patents and patent applications related to CRISPR–Cas systems and uses thereof. H.H.W. is a scientific advisor to SNIPR Biome. S.H.S. is a co-founder and scientific advisor to Dahlia Biosciences and an equity holder in Dahlia Biosciences and Caribou Biosciences.
Additional information
Peer review information Nature Biotechnology thanks Joseph Bondy-Denomy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15.
Supplementary Tables
Supplementary Tables 1–7.
Source data
Source Data Fig. 3
Unprocessed gels
Source Data Fig. 4
Unprocessed gels
Source Data Fig. 5
Unprocessed gels
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vo, P.L.H., Ronda, C., Klompe, S.E. et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat Biotechnol 39, 480–489 (2021). https://doi.org/10.1038/s41587-020-00745-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-00745-y
This article is cited by
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Targeted DNA integration in human cells without double-strand breaks using CRISPR-associated transposases
Nature Biotechnology (2024)
-
Precise integration of large DNA sequences in plant genomes using PrimeRoot editors
Nature Biotechnology (2024)
-
Bacterial genome engineering using CRISPR-associated transposases
Nature Protocols (2024)