Abstract
Targeted saturation mutagenesis of crop genes could be applied to produce genetic variants with improved agronomic performance. However, tools for directed evolution of plant genes, such as error-prone PCR or DNA shuffling, are limited1. We engineered five saturated targeted endogenous mutagenesis editors (STEMEs) that can generate de novo mutations and facilitate directed evolution of plant genes. In rice protoplasts, STEME-1 edited cytosine and adenine at the same target site with C > T efficiency up to 61.61% and simultaneous C > T and A > G efficiency up to 15.10%. STEME-NG, which incorporates the nickase Cas9-NG protospacer-adjacent motif variant, was used with 20 individual single guide RNAs in rice protoplasts to produce near-saturated mutagenesis (73.21%) for a 56-amino-acid portion of the rice acetyl-coenzyme A carboxylase (OsACC). We also applied STEME-1 and STEME-NG for directed evolution of the OsACC gene in rice and obtained herbicide resistance mutations. This set of two STEMEs will accelerate trait development and should work in any plants amenable to CRISPR-based editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available in the article and its supplementary figures and tables or are available from the corresponding author upon request. For sequence data, rice LOC_Os IDs LOC_Os01g55540 (OsAAT), LOC_Os05g22940 (OsACC), LOC_Os03g05730 (OsCDC48), LOC_Os09g26999 (OsDEP1), LOC_Os02g11010 (OsOD, OsEV) are available on the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The deep sequencing data have been deposited with the NCBI BioProject database under accession code numbers PRJNA590652 and PRJNA590653. Plasmids encoding STEME-1, STEME-2, STEME-3, STEME-4, STEME-NG, A3A-PBE-NG, PABE-7-NG, pH-STEME-1-esgRNA and pH-STEME-NG-esgRNA will be available from Addgene.
Code availability
The custom Python script to analyze types of mutational reads and amino acid substitutions is in the Supplementary Code file.
References
Engqvist, M. K. M. & Rabe, K. S. Applications of protein engineering and directed evolution in plant research. Plant Physiol. 179, 907–917 (2019).
Henikoff, S., Till, B. J. & Comai, L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636 (2004).
Slade, A. J., Fuerstenberg, S. I., Loeffler, D., Steine, M. N. & Facciotti, D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23, 75–81 (2005).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Garst, A. D. et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55 (2016).
Halperin, S. O. et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252 (2018).
Bao, Z. et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat. Biotechnol. 36, 505–508 (2018).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Ran, Y., Liang, Z. & Gao, C. Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 60, 490–505 (2017).
Butt, H. et al. CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20, 73 (2019).
Zhang, Y. & Qi, Y. CRISPR enables directed evolution in plants. Genome Biol. 20, 83 (2019).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, 1248 (2016).
Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035 (2016).
Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 13, 1036–1042 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Li, Q. et al. CRISPR–Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 20, 1315–1325 (2018).
Mol, C. D. et al. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell 82, 701–708 (1995).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Zong, Y. et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953 (2018).
Li, Z., Xiong, X. & Li, J.-F. New cytosine base editor for plant genome editing. Sci. China Life Sci. 61, 1602–1603 (2018).
Li, C. et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59 (2018).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Powles, S. B. & Yu, Q. Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347 (2010).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Zhou, C. et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278 (2019).
Grünewald, J. et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 37, 1041–1048 (2019).
Zhang, H., Yang, Z., Shen, Y. & Tong, L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 299, 2064–2067 (2003).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Shan, Q. et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 6, 1365–1368 (2013).
Meng, X. et al. Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 10, 1238–1241 (2017).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Zhang, H., Tweel, B. & Tong, L. Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme-A carboxylase by haloxyfop and diclofop. Proc. Natl Acad. Sci. USA 101, 5910–5915 (2004).
Xiang, S., Callaghan, M. M., Watson, K. G. & Tong, L. A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim. Proc. Natl Acad. Sci. USA 106, 20723–20727 (2009).
Yu, L. P., Kim, Y. S. & Tong, L. Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden. Proc. Natl Acad. Sci. USA 107, 22072–22077 (2010).
Eswar, N. et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31, 3375–3380 (2003).
Bae, S., Park, J. & Kim, J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Acknowledgements
We thank S. Jin for technical support on bioinformatic analysis. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, grant no. XDA24000000), the National Natural Science Foundation of China (grant nos. 31788103 and 31900301) and the National Key Research and Development Program of China (grant no. 2016YFD0101804).
Author information
Authors and Affiliations
Contributions
C. Li, R.Z., X.M., J.L. and C.G. designed the project. C. Li, R.Z., S.C., Y.Z. and C. Lu performed most of the experiments. X.M. performed transformation. Y.-H.C. analyzed the three-dimensional structure of the CT domain. J.L. and C.G. supervised the project. C. Li, R.Z., J.-L.Q., Y.-H.C., J.L. and C.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have submitted a patent application based on the results reported in this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Activities and product purity of STEME-1 to STEME-4 in rice protoplasts.
(a)-(f) are OsAAT, OsACC, OsCDC48, OsDEP1, OsEV, and OsOD targets, respectively. One of three independent experiments is shown.
Supplementary Fig. 2 The allelic outcomes and indel value of STEMEs in rice protoplasts.
(a) The allelic outcomes of rice sites edited by STEME-1 (n=1 shown) in rice protoplasts. The simultaneous C>T and A>G editing efficiency of STEME-1 was ranged from 0.68-15.37% vs. 0.69-15.50% while only count the A>G substitutions (including single A>G substitution and simultaneous C>T and A>G substitutions) (Fig. 1d–f). (b) Indel frequencies of STEMEs and Cas9 in rice protoplasts. Comparison of indel frequencies by the A3A-PBE, PABE-7, STEME-1, STEME-2, STEME-3, and STEME-4 constructs in rice protoplasts. Values and error bars indicate the mean ± s.e.m of three independent experiments.
Supplementary Fig. 3 An overview of NG PAM targets at different rice loci.
(a) Architectures of the A3A-PBE-NG, PABE7-NG and pCas9-NG. ecTadA7.10, evolved Escherichia coli TadA; aa, amino acid; XTEN, a 16-aa linker; NLS, nuclear localization signal; CaMV, cauliflower mosaic virus; Term, terminator. (b) Sequences of 16 NG PAM targets in four loci of the rice genome. The PAM sequences are shown in red. (c) Diagram of OsAAT, OsCDC48, OsDEP1 and OsODEV, and their NG PAM targets. Regions flanking about 80bp of the OsAAT, OsCDC48, OsDEP1 and OsODEV loci were selected to design 20-nt sgRNA sequences to reduce the possible influence of chromatin state on editing.
Supplementary Fig. 4 Activities and product purity of STEME-NG in rice protoplasts.
(a)-(p) are NG PAM targets in OsAAT, OsCDC48, OsDEP1, and OsODEV, respectively. One of three independent experiments is shown.
Supplementary Fig. 5 Base editing efficiencies by A3A-PBE-NG and PABE7-NG in rice protoplasts.
Both A3A-PBE-NG (a) and PABE7-NG (b) had broad capacities for editing the targets of NGA, NGT, NGC or NGG PAMs. An untreated protoplast sample served as control. Values and error bars indicate the mean ± s.e.m of three independent experiments.
Supplementary Fig. 6 Indel frequencies of A3A-PBE-NG, PABE7-NG and STEME-NG compared with Cas9-NG in rice protoplasts.
A3A-PBE, PABE-7, STEME-1 and Cas9 together with NGG PAM sgRNAs served as positive control. The resulting efficiencies were log scaled by the base number 10. An untreated protoplast sample served as a negative control. Values and error bars indicate the mean ± s.e.m of three independent experiments.
Supplementary Fig. 7 Saturated mutagenesis of a 168-bp region of OsACC by A3A-PBE-NG in rice protoplasts.
A3A-PBE-NG (a) and the untreated control (b) are shown. Max values were calculated when different targets converted the same cytosine and guanine. Values and error bars indicate the mean ± s.e.m of three independent experiments.
Supplementary Fig. 8 C>T and A>G simultaneous editing and indel frequencies of 20 OsACC targets in rice protoplasts.
(a) The simultaneous editing frequencies of C>T and A>G in 20 targets of OsACC by STEME-NG in rice protoplasts. (b) Indel frequencies of 20 OsACC targets by A3A-PBE-NG, STEME-NG and Cas9-NG in rice protoplasts. An untreated protoplast sample served as a negative control. Values and error bars indicate the mean ± s.e.m of three independent experiments.
Supplementary Fig. 9 Saturated substitutions of 56 amino acids in the OsACC CT domain by A3A-PBE-NG in rice protoplasts.
Frequencies of silent, missense, and nonsense mutations were collected.
Supplementary Fig. 10 An overview of sgRNAs for saturated substituting 400 amino acids in the OsACC CT domain.
(a) An overview of the saturated targets on OsACC CT domain for rice plants. (b) Architectures of the binary vectors pH-STEME-1-esgRNA and pH-STEME-NG-esgRNA. ecTadA7.10, evolved Escherichia coli TadA; aa, amino acid; XTEN, a 16-aa linker; NLS, nuclear localization signal; E9 Term, pea rbcS-E9 terminator; pd35S, cauliflower mosaic virus double 35S promoter; Hyg, hygromycin. Arrows indicate the primers for amplicon deep sequencing.
Supplementary Fig. 11 Base editing and indel frequencies by STEMEs in rice plants.
(a) Base editing efficiencies of STEMEs in each transformed group. (b) Indel efficiencies of STEMEs in each transformed group. An average 0.90% indel reads in these groups, and 31.47% of the indels conferred in-frame mutations (Supplementary Dataset 2). Wild-type plants were used as untreated controls.
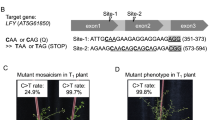
Supplementary Fig. 12 Genotypes of the P1927F, W2125C, S1866F, and A1884P mutants.
(a) The P1927F and Q1926* changes were caused by C:G>T:A transitions. Asterisk represents the stop codon. (b) The A2123T and W2125C changes were caused by a C:G>T:A transition and a C:G>G:C transversion, respectively. (c) The S1866F change was caused by a C:G>T:A transition and a silent mutation of G1869 was the result of a C:G>T:A transition. (d) The A1884P change was caused by a C:G>G:C transversion (A1884P-Hetero1) or simutaniously C:G>G:C transversion and A:T>G:C transition (A1884P-Hetero2). The edited bases are shown in lower case. Red triangles indicate peaks of edited bases. The genotypes of these mutations with two double peaks were confirmed further by T-vector cloning.
Supplementary Fig. 13 Selecting and genotyping haloxyfop resistance seedlings.
(a) Schematic of the procedure for mutating the OsACC CT domain via STEMEs on medium supplemented with haloxyfop using Groups 6 and 20 of individual sgRNAs. (b-c) The percentage of sgRNA sequencing reads after selection on medium containing 0.108 mg l-1 haloxyfop for 4 weeks in Group 6 (b) and Group 20 (c). The sgRNA OsACC-sg33 covered P1927 in Group 6 accounts for 100%, OsACC-sg140 and OsACC-sg141 both covered W2125 in Group 20 account for 96.74%. For the mutational reads of target region, 63.53% mutated sequences involved P1927F in Group 6, 48.71% mutated sequences involved W2125C in Group 20 (Supplementary Dataset 3). (d) Herbicide resistance seedlings harboring P1927F and W2125C mutations, respectively, regenerated in haloxyfop-containing medium. One biological experiment was performed. Scale bar, 2 cm. (e) Genotypes of herbicide resistance seedlings from Group 6 and Group 20. The P1927F and Q1926* were caused by C:G>T:A transitions; the A2123T was caused by C:G>T:A transition or C:G>T:A and A:T>G:C simultaneously conversions; the W2125C changes were caused by a C:G>G:C transversion. The edited bases are shown in lower case. Asterisk represents the stop codon. Red triangles indicate peaks of edited bases. The genotypes of these mutations with two double peaks were confirmed further by T-vector cloning.
Supplementary Fig. 14 Sequencing of OsACC full-length genomic DNA of P1927F, W2125C, S1866F, and A1884P mutants.
Twenty-two paired primers were used to sequence OsACC genomic DNA. The base changes of each mutant in P1927F, W2125C, S1866F, and A1884P codons were indicated. No other mutational changes occurred except for these target sites.
Supplementary Fig. 15 Phenotypes and genotypes of two A2123T mutants.
(a) Phenotypes of two homozygous A2123T mutants. One biological experiment was performed. Scale bar, 2 cm. Red arrows indicate the died symptom of newly grown leaves. (b) Genotypes of A2123T mutants. The A2123T change was caused by a C:G>T:A transition. The edited bases are shown in lower case. Red triangles indicate peaks of edited bases.
Supplementary information
Supplementary Materials
Supplementary Figs. 1–15, Supplementary Tables 1–8, Supplementary Sequences 1–3 and Supplementary Code.
Supplementary Dataset 1
Details of the saturated mutagenesis of 56 amino acids of the OsACC CT domain in rice protoplasts.
Supplementary Dataset 2
Mutational reads of directed evolution of 400 amino acids of the OsACC CT domain in rice plants.
Supplementary Dataset 3
Mutational reads of using STEMEs for targeted mutagenesis under concurrent selection pressure.
Rights and permissions
About this article
Cite this article
Li, C., Zhang, R., Meng, X. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38, 875–882 (2020). https://doi.org/10.1038/s41587-019-0393-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0393-7
This article is cited by
-
Precise fine-turning of GhTFL1 by base editing tools defines ideal cotton plant architecture
Genome Biology (2024)
-
Application of genome editing techniques to regulate gene expression in crops
BMC Plant Biology (2024)
-
CRISPR/Cas-mediated germplasm improvement and new strategies for crop protection
Crop Health (2024)
-
Directed mutagenesis in plants through genome editing using guide RNA library
The Nucleus (2024)
-
Fusion of a rice endogenous N-methylpurine DNA glycosylase to a plant adenine base transition editor ABE8e enables A-to-K base editing in rice plants
aBIOTECH (2024)