Abstract
The Sleeping Beauty (SB) transposon system is an efficient non-viral gene transfer tool in mammalian cells, but its broad use has been hampered by uncontrolled transposase gene activity from DNA vectors, posing a risk of genome instability, and by the inability to use the transposase protein directly. In this study, we used rational protein design based on the crystal structure of the hyperactive SB100X variant to create an SB transposase (high-solubility SB, hsSB) with enhanced solubility and stability. We demonstrate that hsSB can be delivered with transposon DNA to genetically modify cell lines and embryonic, hematopoietic and induced pluripotent stem cells (iPSCs), overcoming uncontrolled transposase activity. We used hsSB to generate chimeric antigen receptor (CAR) T cells, which exhibit potent antitumor activity in vitro and in xenograft mice. We found that hsSB spontaneously penetrates cells, enabling modification of iPSCs and generation of CAR T cells without the use of transfection reagents. Titration of hsSB to modulate genomic integration frequency achieved as few as two integrations per genome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data generated or analyzed during this study and all unique materials can be made available by the corresponding authors upon reasonable request.
References
Turtle, C. J. et al. Durable molecular remissions in chronic lymphocytic leukemia treated with CD19-specific chimeric antigen receptor-modified T cells after failure of ibrutinib. J. Clin. Oncol. 35, 3010–3020 (2017).
Klebanoff, C. A., Rosenberg, S. A. & Restifo, N. P. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat. Med. 22, 26–36 (2016).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Lamers, C. H. et al. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood 117, 72–82 (2011).
Bushman, F. et al. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3, 848–858 (2005).
Clauss, J. et al. Efficient non-viral T-cell engineering by Sleeping Beauty minicircles diminishing DNA toxicity and miRNAs silencing the endogenous T-cell receptors. Hum. Gene Ther. 29, 569–584 (2018).
Cornu, T. I., Mussolino, C. & Cathomen, T. Refining strategies to translate genome editing to the clinic. Nat. Med. 23, 415–423 (2017).
Li, K., Wang, G., Andersen, T., Zhou, P. & Pu, W. T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PloS One 9, e105779 (2014).
Peng, P. D. et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 16, 1042–1049 (2009).
Ivics, Z. et al. Transposon-mediated genome manipulation in vertebrates. Nat. Methods 6, 415–422 (2009).
Tipanee, J., Chai, Y. C., VandenDriessche, T. & Chuah, M. K. Preclinical and clinical advances in transposon-based gene therapy. Biosci Rep. 37, BSR20160614 (2017).
Vargas, J. E. et al. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 14, 288 (2016).
Klompe, S. E., Vo, P. L. H., Halpin-Healy, T. S. & Sternberg, S. H. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019).
Morero, N. R. et al. Targeting IS608 transposon integration to highly specific sequences by structure-based transposon engineering. Nucleic Acids Res. 46, 4152–4163 (2018).
Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019).
Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997).
Dupuy, A. J., Jenkins, N. A. & Copeland, N. G. Sleeping beauty: a novel cancer gene discovery tool. Hum. Mol. Genet. 15 (Spec. 1), R75–R79 (2006).
Hodge, R., Narayanavari, S. A., Izsvak, Z. & Ivics, Z. Wide awake and ready to move: 20 years of non-viral therapeutic genome engineering with the Sleeping Beauty transposon system. Hum. Gene Ther. 28, 842–855 (2017).
Hudecek, M. et al. Going non-viral: the Sleeping Beauty transposon system breaks on through to the clinical side. Crit. Rev. Biochem. Mol. Biol. 52, 355–380 (2017).
Kawakami, K., Largaespada, D. A. & Ivics, Z. Transposons as tools for functional genomics in vertebrate models. Trends Genet. 33, 784–801 (2017).
Monjezi, R. et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 31, 186–194 (2017).
Kebriaei, P. et al. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 126, 3363–3376 (2016).
Wang, Z. et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 11, 711–721 (2004).
Galla, M. et al. Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res. 39, 7147–7160 (2011).
Huang, X. et al. Unexpectedly high copy number of random integration but low frequency of persistent expression of the Sleeping Beauty transposase after trans delivery in primary human T cells. Hum. Gene Ther. 21, 1577–1590 (2010).
Liang, Q., Kong, J., Stalker, J. & Bradley, A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47, 404–408 (2009).
Wilber, A. et al. Messenger RNA as a source of transposase for Sleeping Beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 15, 1280–1287 (2007).
Holstein, M. et al. Efficient non-viral gene delivery into human hematopoietic stem cells by minicircle Sleeping Beauty transposon vectors. Mol. Ther. 26, 1137–1153 (2018).
Hackett, P. B., Starr, T. K. & Cooper, L. J. N. in Translating Gene Therapy to the Clinic (eds. J. Laurence & M. Franklin) 65–83 (Academic, 2015).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Gaj, T., Guo, J., Kato, Y., Sirk, S. J. & Barbas, C. F. 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods 9, 805–807 (2012).
Zayed, H., Izsvak, Z., Khare, D., Heinemann, U. & Ivics, Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31, 2313–2322 (2003).
Paatero, A. O. et al. Bacteriophage Mu integration in yeast and mammalian genomes. Nucleic Acids Res. 36, e148 (2008).
Trubitsyna, M. et al. Use of mariner transposases for one-step delivery and integration of DNA in prokaryotes and eukaryotes by transfection. Nucleic Acids Res. 45, e89 (2017).
Voigt, F. et al. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat. Commun. 7, 11126 (2016).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Mates, L. et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 (2009).
Avramopoulou, V., Mamalaki, A. & Tzartos, S. J. Soluble, oligomeric, and ligand-binding extracellular domain of the human α7 acetylcholine receptor expressed in yeast: replacement of the hydrophobic cysteine loop by the hydrophilic loop of the ACh-binding protein enhances protein solubility. J. Biol. Chem. 279, 38287–38293 (2004).
Slusarczyk, H., Felber, S., Kula, M. R. & Pohl, M. Stabilization of NAD-dependent formate dehydrogenase from Candida boidinii by site-directed mutagenesis of cysteine residues. Eur. J. Biochem. 267, 1280–1289 (2000).
Zhao, Y., Stepto, H. & Schneider, C. K. Development of the first world health organization lentiviral vector standard: toward the production control and standardization of lentivirus-based gene therapy products. Hum. Gene Ther. Methods 28, 205–214 (2017).
Davila, M. L. et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 6, 224ra225 (2014).
Maude, S. L. et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014).
Lee, D. W. et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015).
Wang, X. et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263 (2011).
Ivics, Z., Izsvak, Z., Minter, A. & Hackett, P. B. Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl Acad. Sci. USA 93, 5008–5013 (1996).
Charrier, S. et al. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther. 18, 479–487 (2011).
Luo, G., Ivics, Z., Izsvak, Z. & Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 95, 10769–10773 (1998).
Hackett, P. B., Largaespada, D. A., Switzer, K. C. & Cooper, L. J. Evaluating risks of insertional mutagenesis by DNA transposons in gene therapy. Transl. Res. 161, 265–283 (2013).
Chusainow, J. et al. A study of monoclonal antibody-producing CHO cell lines: what makes a stable high producer? Biotechnol. Bioeng. 102, 1182–1196 (2009).
Lu, T. L. et al. A rapid cell expansion process for production of engineered autologous CAR-T cell therapies. Hum. Gene Ther. Methods 27, 209–218 (2016).
Gattinoni, L. et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011).
Klebanoff, C. A., Gattinoni, L. & Restifo, N. P. Sorting through subsets: which T-cell populations mediate highly effective adoptive immunotherapy? J. Immunother. 35, 651–660 (2012).
Turtle, C. J. et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 126, 2123–2138 (2016).
Maus, M. V. et al. T cells expressing chimeric antigen receptors can cause anaphylaxis in humans. Cancer Immunol. Res. 1, 26–31 (2013).
D’Aloia, M. M., Zizzari, I. G., Sacchetti, B., Pierelli, L. & Alimandi, M. CAR-T cells: the long and winding road to solid tumors. Cell Death Dis. 9, 282 (2018).
Roth, T. L. et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Bhatt, S. & Chalmers, R. Targeted DNA transposition in vitro using a dCas9–transposase fusion protein. Nucleic Acids Res. 47, 8126–8135 (2019).
Hew, B. E., Sato, R., Mauro, D., Stoytchev, I. & Owens, J. B. RNA-guided piggyBac transposition in human cells. Synth. Biol. (Oxf). 4, ysz018 (2019).
Voigt, K. et al. Retargeting Sleeping Beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol. Ther. 20, 1852–1862 (2012).
Wang, M., Glass, Z. A. & Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 24, 144–150 (2017).
Makarova, O., Kamberov, E. & Margolis, B. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29, 970–972 (2000
Ruf, S. et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat. Genet. 43, 379–386 (2011).
Hudecek, M. et al. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol. Res. 3, 125–135 (2015).
Hudecek, M. et al. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood 116, 4532–4541 (2010).
Hudecek, M. et al. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin. Cancer Res. 19, 3153–3164 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Chiang, C. W. et al. Ultraconserved elements in the human genome: association and transmission analyses of highly constrained single-nucleotide polymorphisms. Genetics 192, 253–266 (2012).
Acknowledgements
The authors thank F. Dyda, K.R. Patil, M. Beck and members of the Barabas lab for helpful discussions and the Nature Editing Service for editing assistance. We also thank the Flow Cytometry Core Facility, the Genomics Core Facility, the Advanced Light Microscopy Facility and the Protein Expression and Purification Core Facility at EMBL Heidelberg for materials and support. We thank Z. Izsvak (Max Delbrück Center) for providing the CMV(CAT)T7-SB100X plasmid and F. Spitz (EMBL Heidelberg) for the pT2/PGK-neo construct; A. Aulehla for providing access to the Neon transfection system and reagents; S. Henkel for providing reagents and assistance for mESC culture and immunostaining; H. Bönig (Blutspendedienst des Deutschen Roten Kreuz) for providing human HSPCs; K.-M. Noh and N. Diaz for providing materials and advice with iPSC culturing; V. Rybin for assistance and advice with the CD spectroscopy experiments; and V. Benes for assistance and support with sequence analysis. This work was supported by the EMBL, the Paul Ehrlich Institute, the EMBL International PhD Programme (fellowship to I.Q.), the Deutsche Forschungsgemeinschaft (project number 324392634, TRR 221 to M.H. and H.E.) and German Cancer Aid (Deutsche Krebshilfe, Max Eder Program Award 70110313 to M.H.). M.H. was supported by the Young Scholar Program of the Bavarian Academy of Sciences (Junges Kolleg, Bayerische Akademie der Wissenschaften) and the m4 Award in Personalized Medicine (Free State of Bavaria, BIO-1601-0002). Z.I. was supported by the Center for Cell and Gene Therapy of the LOEWE (Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz) program in Hessen, Germany, and by a grant from the Deutsche Forschungsgemeinschaft (IV 21/11-1).
Author information
Authors and Affiliations
Contributions
I.Q., A.M., C.Z., M.H. and O.B. designed the research. All authors contributed to analysis and discussion of the results and read and approved the manuscript. I.Q., A.M., C.Z., M.H. and O.B. wrote the manuscript with input from all authors. I.Q. designed, screened and characterized transposase variants. A.M., M.A. and M.M. performed T cell engineering and characterization. C.Z. conducted transposition assays and engineering of HeLa, CHO, mESC and iPSC lines. C.M. performed integration site profiling in T cells and copy number analyses. E.G. performed HSPC engineering. T.R. analyzed integration sites in HeLa cells. H.E. provided expert advice and support during the project. Z.I. supervised integration profiling, copy number analyses and HSPC engineering and participated in coordinating the research. M.H. supervised T cell engineering and analyses and oversaw the project. O.B. supervised protein work and cell line engineering and oversaw the project.
Corresponding authors
Ethics declarations
Competing interests
Z.I. is an inventor on patents concerning the development and use of the Sleeping Beauty technology (proprietor Max-Delbrück-Centrum für Molekulare Medizin; patents US9228180B2 and EP2160461B1), and two patent applications have been filed with the European Patent Office concerning the hsSB transposase (EP17187128 and EP19158066.1).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Biochemical characterization of the hsSB transposase.
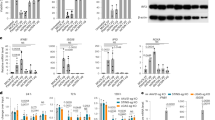
a, SDS-PAGE analysis of samples collected during recombinant production and purification of the hsSB transposase variant. hsSB was produced in E. coli (fused to N-terminal purification and solubility tags) at high yields and was highly pure (>95%) after tag removal and size exclusion chromatography. b, SDS-PAGE analysis of purified SB proteins after concentration (conc.). hsSB can be concentrated up to 50 fold (corresponding to 20 mg per ml), whereas SB100X undergoes precipitation at concentrations higher than 7 mg per ml. The vast majority of hsSB remains in the soluble fraction in the low-salt R buffer used for electroporation, even at high protein concentration. c, CD spectra of the SB100X (blue curve) and hsSB (red curve) proteins. a–c, Experiments were repeated independently two b and three a,c times with similar results.
Supplementary Figure 2 Transposition assays in HeLa cells.
a, Representative transposition assay. Transgene (neor) insertions generated G418-resistant colonies after hsSB transposase transfection. Experiments were repeated independently three times with similar results. b, Number of resistant colonies with various amounts of hsSB provided on a plasmid vector. Mean values (n = 2 independent experiments).
Supplementary Figure 3
Retention time of hsSB delivered into HeLa cells as a protein or expressed from plasmid DNA. Western blot analysis was performed on lysates from HeLa cells transfected with 0.5 µg of transposon DNA (pT2/PGK-neo) and electroporated with 10 μg of the hsSB protein or transfected with 500 ng of the hsSB expression plasmid. Samples were collected at the indicated time points, and 20 µg of the total cell lysate was separated by electrophoresis and transferred to a nitrocellulose membrane. The SB transposase was detected with an anti-SB antibody. The internal loading control was GAPDH, detected with an anti-GAPDH antibody. a, Full scan of the blots shown in Fig. 2d. b, Measurement of the intensities of the bands allowed for the quantification of hsSB persistence in HeLa cells over time. Experiments were repeated independently two times with similar results.
Supplementary Figure 4 Transposition assays in mESCs.
a, Representative transposition assay. Transgene (neor) insertions generated G418-resistant colonies after transfection of increasing amounts of hsSB transposase. Transposition assay cultures were diluted 1:10 for plating. Colony counts were corrected for the dilution and plotted. b, Flow cytometry plots showing the percentage of cells expressing the Oct4 pluripotency marker versus the Venus-positive cells. a,b, Experiments were performed once.
Supplementary Figure 5 Characterization of CAR T cell products.
a, Cytokine secretion of CAR T cells produced by transfection with CD19 CAR MC and hsSB protein (MC-hsSB) or CD19 CAR MC and SB100X MC (MC-MC). The concentrations of IL-2 and IFN-γ were measured by ELISA in cell culture media after a 24-h co-culture with target cells (E:T = 4:1). b, CAR T cells were labelled with carboxyfluorescein succinimidyl ester and co-cultured with irradiated target cells (E:T = 4:1). Proliferation was measured by the CFSE dilution of CAR T cells after 72 h (NT = non-transfected). a,b, Data were obtained from n = 3 independent experiments with n = 3 different T cell donors. Mean values; error bars represent s.d. c, Consensus sequence of SB insertion sites in T cells taken from the batch used for injection into the Raji xenograft mouse model. d, Heatmap demonstrating the insertion frequencies into genes of various expression levels (expr.lev).
Supplementary Figure 6 Supplementary data from the Raji xenograft mouse model.
a, Ventral and dorsal bioluminescence images of mice treated with CD19 CAR T cells or non-transfected (NT) control T cells (as in Fig. 4) taken at the indicated time points. b, Group analysis of tumor growth. Mean values; error bars represent s.d. Group sizes are identical to the images displayed in a: n = 2, 3, 5 and 5 animals for untreated, NT, MC-MC and MC-hsSB samples, respectively. c, Kaplan–Meier survival curves.
Supplementary Figure 7 Full scans of the Western blots analyzing hsSB uptake.
a, Blots showing cellular uptake and retention of hsSB in HeLa cells upon addition to the culture medium shown in Fig. 5c. The SB transposase was detected with anti-SB antibody. The internal loading control was GAPDH, blotted with an anti-GAPDH antibody. b, Blots for hsSB penetration from the culture media in iPSCs, blotted with anti-SB or anti-lamin B1/B2 (loading control) antibody as shown in Fig. 5e.
Supplementary Figure 8 Characterization of T cells after spontaneous hsSB penetration.
a, hsSB penetration in stimulated and non-stimulated CD4+ T cells. Immunofluorescence imaging of T cells showing DAPI-stained nuclei (blue), hsSB staining (green) and the merge. Cells stained in the absence of primary SB antibody are shown below (IF control). Experiments were repeated two times using cells from different donors with similar results. b, Functional analysis of CD19 CAR T cells generated with spontaneous hsSB penetration (MC-hsSB) or by transfection of SB100X MC (MC-MC). Cytokine secretion of CAR T cells. CD8+ T cells from healthy donors were transfected with CD19 CAR MC, incubated with hsSB, enriched for CAR-modified cells (EGFRt positive) and expanded with irradiated CD19+ B-LCL cells before functional assays. Results from one experiment with one T cell donor are shown. The concentrations of IL-2 and IFN-γ were measured by ELISA in cell culture media after a 24-h co-culture with target cells (E:T = 4:1). Mean values; error bars represent s.d. (n = 3 independent measurements with 1 T cell donor). NT = non-transfected.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Supplementary Tables 1–3
Rights and permissions
About this article
Cite this article
Querques, I., Mades, A., Zuliani, C. et al. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat Biotechnol 37, 1502–1512 (2019). https://doi.org/10.1038/s41587-019-0291-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0291-z
This article is cited by
-
Targeted DNA integration in human cells without double-strand breaks using CRISPR-associated transposases
Nature Biotechnology (2024)
-
AAV-mediated delivery of a Sleeping Beauty transposon and an mRNA-encoded transposase for the engineering of therapeutic immune cells
Nature Biomedical Engineering (2023)
-
Construction and Quantitative Evaluation of a Tissue-Specific Sleeping Beauty by EDL2-Specific Transposase Expression in Esophageal Squamous Carcinoma Cell Line KYSE-30
Molecular Biotechnology (2023)
-
A novel aptamer beacon for rapid screening of recombinant cells and in vivo monitoring of recombinant proteins
Applied Microbiology and Biotechnology (2023)
-
Targeted delivery of a PD-1-blocking scFv by CD133-specific CAR-T cells using nonviral Sleeping Beauty transposition shows enhanced antitumour efficacy for advanced hepatocellular carcinoma
BMC Medicine (2023)