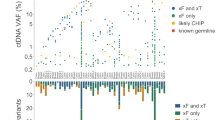
Abstract
Genomic analysis of paired tumor–normal samples and clinical data can be used to match patients to cancer therapies or clinical trials. We analyzed 500 patient samples across diverse tumor types using the Tempus xT platform by DNA-seq, RNA-seq and immunological biomarkers. The use of a tumor and germline dataset led to substantial improvements in mutation identification and a reduction in false-positive rates. RNA-seq enhanced gene fusion detection and cancer type classifications. With DNA-seq alone, 29.6% of patients matched to precision therapies supported by high levels of evidence or by well-powered studies. This proportion increased to 43.4% with the addition of RNA-seq and immunotherapy biomarker results. Combining these data with clinical criteria, 76.8% of patients were matched to at least one relevant clinical trial on the basis of biomarkers measured by the xT assay. These results indicate that extensive molecular profiling combined with clinical data identifies personalized therapies and clinical trials for a large proportion of patients with cancer and that paired tumor–normal plus transcriptome sequencing outperforms tumor-only DNA panel testing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
VCF files, RNA count files and associated deidentified clinical data that support these findings will be available through Vivli (ID T19.01).
References
Fernandes, G. et al. Next-generation sequencing-based genomic profiling: ostering innovation in cancer care? Clinics 72, 588–594 (2017).
Radovich, M. et al. Clinical benefit of a precision medicine based approach for guiding treatment of refractory cancers. Oncotarget 7, 56491–56500 (2016).
Dhir, M. et al. Impact of genomic profiling on the treatment and outcomes of patients with advanced gastrointestinal malignancies. Cancer Med. 6, 195–206 (2017).
Wheler, J. J. et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res. 76, 3690–3701 (2016).
Gong, J. et al. Value-based genomics. Oncotarget 9, 15792–15815 (2018).
The ASCO Post. 2018 ASCO: IMPACT trial matches treatment to genetic changes in the tumor to improve survival across multiple cancer types.The ASCO Post http://www.ascopost.com/News/58897 (2 June 2018).
Beaubier, N. et al. Clinical validation of the tempus xT next-generation sequencing targeted oncology assay. Oncotarget 10, 2384–2396 (2019).
Goodman, A. M. et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 (2017).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Miller, A. et al. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 7, e612 (2017).
Desrichard, A., Snyder, A. & Chan, T. A. Cancer neoantigens and applications for immunotherapy. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-14-3175 (2016).
Reiman, D. et al. Integrating RNA expression and visual features for immune infiltrate prediction. Biocomputing 2019, 284–295 (2018).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Newton, Y. et al. TumorMap: exploring the molecular similarities of cancer samples in an interactive portal. Cancer Res. 77, e111–e114 (2017).
Solomon, B., Varella-Garcia, M. & Camidge, D. R. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J. Thorac. Oncol. 4, 1450–1454 (2009).
Chae, Y. K. et al. Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-small cell lung cancer. Clin. Lung Cancer https://doi.org/10.1016/J.CLLC.2018.09.008 (2018).
Rooney, M. S. et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Roufas, C. et al. The expression and prognostic impact of immune cytolytic activity-related markers in human malignancies: a comprehensive meta-analysis. Front. Oncol. 8, 27 (2018).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Li, M. M. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J. Mol. Diagnostics 19, 4–23 (2017).
Wang, Z. et al. Significance of the TMPRSS2:ERG gene fusion in prostate cancer. Mol. Med. Rep. 16, 5450–5458 (2017).
Chatterjee, P. et al. The TMPRSS2-ERG gene fusion blocks XRCC4-mediated nonhomologous end-joining repair and radiosensitizes prostate cancer cells to PARP inhibition. Mol. Cancer Ther. 14, 1896–1906 (2015).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Hegde, G. V. et al. Blocking NRG1 and other ligand-mediated Her4 signaling enhances the magnitude and duration of the chemotherapeutic response of non-small cell lung cancer. Sci. Transl. Med. 5, 171ra18 (2013).
Sheng, Q. et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell 17, 298–310 (2010).
Han, M.-E. et al. Overexpression of NRG1 promotes progression of gastric cancer by regulating the self-renewal of cancer stem cells. J. Gastroenterol. 50, 645–656 (2015).
Yun, S. et al. Clinical significance of overexpression of NRG1 and its receptors, HER3 and HER4, in gastric cancer patients. Gastric Cancer 21, 225–236 (2018).
Luraghi, P. et al. A molecularly annotated model of patient-derived colon cancer stem-like cells to assess genetic and nongenetic mechanisms of resistance to anti-EGFR therapy. Clin. Cancer Res. 24, 807–820 (2018).
Yonesaka, K. et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci. Transl. Med. 3, 99ra86 (2011).
Yang, L. et al. NRG1-dependent activation of HER3 induces primary resistance to trastuzumab in HER2-overexpressing breast cancer cells. Int. J. Oncol. 51, 1553–1562 (2017).
Wilson, T. R., Lee, D. Y., Berry, L., Shames, D. S. & Settleman, J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell 20, 158–172 (2011).
Mendell, J. et al. Clinical translation and validation of a predictive biomarker for patritumab, an anti-human epidermal growth factor receptor 3 (HER3) monoclonal antibody, in patients with advanced non-small cell lung cancer. EBioMedicine 2, 264–271 (2015).
Conway, J. R., Lex, A., Gehlenborg, N. & Hancock, J. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Teer, J. K. et al. Evaluating somatic tumor mutation detection without matched normal samples. Hum. Genomics 11, 22 (2017).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831 (2017).
Hartmaier, R. J. et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 77, 2464–2475 (2017).
Maxwell, K. N. et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 8, 319 (2017).
Yan, M. et al. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev. 34, 157–164 (2015).
Darvin, P., Toor, S. M., Sasidharan Nair, V. & Elkord, E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 50, 165 (2018).
Lau, D., Bobe, A. M. & Khan, A. A. RNA sequencing of the tumor microenvironment in precision cancer immunotherapy. Trends Cancer 5, 149–156 (2019).
Cristescu, R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 362, eaar3593 (2018).
Allen, J. et al. Barriers to patient enrollment in therapeutic clinical trials for cancer: a landscape report. J. Oncol. Navig. Surviv. 9 (2018).
Unger, J. M., Vaidya, R., Hershman, D. L., Minasian, L. M. & Fleury, M. E. Systematic review and meta-analysis of the magnitude of structural, clinical, and physician and patient barriers to cancer clinical trial participation. J. Natl Cancer Inst. 111, 245–255 (2019).
Institute of Medicine. Transforming Clinical Research in the United States: Challenges and Opportunities: Workshop Summary (National Academies Press, 2010); https://doi.org/10.17226/12900
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Faust, G. G. & Hall, I. M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014).
Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Lonsdale, J. et al. The Genotype–Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Peng, L. et al. Large-scale RNA-Seq transcriptome analysis of 4043 cancers and 548 normal tissue controls across 12 TCGA cancer types. Sci. Rep. 5, 13413 (2015).
Goldman, M. et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. Preprint at https://doi.org/10.1101/326470 (2019).
Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Forbes, S. A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Griffith, M. et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, eaag1166 (2017).
Madhavan, S. et al. ClinGen Cancer Somatic Working Group—standardizing and democratizing access to cancer molecular diagnostic data to drive translational research. Pac. Symp. Biocomput. 23, 247–258 (2018).
Dienstmann, R. et al. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol. Oncol. 8, 859–873 (2014).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics And Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Acknowledgements
We are thankful to the operations, product, engineering and clinical data teams at Tempus Labs, including but not limited to U. Pipic, C. Schwalbach, S. Hynes, K. Stenglein, L. Sachse, A. Hoyer, S. Carsanaro, H, Lefkofsky, R. Chang, M. Barber, R. Pe Benito, R. Star, H. Whipple and D. King. We thank the pathology and lab teams for sample processing and data collection. We are grateful to M. Salazar for managing the work required for this manuscript. We thank G. Palmer and A. Schwarzbach for review of the manuscript, M. Kase and A. Hoffman-Peterson for proofreading, and A. Sheals and B. Santacaterina for help with figure aesthetics and assembly. We thank E. Lefkofsky for his support and discussions.
Author information
Authors and Affiliations
Contributions
N.B., M.B., R.H., C.I., R.T. and D.L. led data analysis and interpretation, and wrote sections of the manuscript. N.B. and T.T. performed the pathologic review of the cohort and wrote sections of the manuscript. C.I., J.M., B.D.L., K.P.S., T.T. and N.B. contributed to gene expression and cancer type predictor analyses and figures. D.L., A.L.C., J.F.P., A.L. and A.A.K. contributed to immune analyses and figures. R.T., S.B., J.P. and W.Z. contributed to mutational and genomic rearrangement analyses and figures. R.H., R.T., D.C.H., N.B., A.S. and M.B. contributed to tumor-only and tumor–normal analyses and figures. R.H., N.B., E.K. and M.B. contributed to therapeutic evidence and clinical trial matching analyses. A.M.B. provided critical review of drafts and figures, wrote sections of the manuscript and reviewed the final manuscript. K.P.W. oversaw manuscript preparation, provided scientific direction, wrote sections of the manuscript and reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Authors are employees of Tempus Labs, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and Supplementary Tables 1–3
Supplementary Table 4
Identification of clinical trial options. Examples of the clinical data fields used to identify pertinent clinical trials for the cohort (n = 481 patients). Multiple clinical trial options may have been reported, but only one is shown per patient in the table.
Supplementary Table 5
Tumor-only analysis of somatic false positives. List of variants classified as somatic and verified as germline. Each variant contains the hg19 coordinates followed by symbol, variant, allele frequency (AF) in the tumor and germline sample, and mutation classification (TVUS, tumor-only variant of unknown significance; TMUT, tumor-only mutation).
Supplementary Table 6
Comparison between full Tempus xT test and tumor-only tests. Comparison of test results and relevant therapies for 50 patients from a full Tempus xT test, a tumor-only DNA sequencing xT test, and an analysis of treatment options based on tumor-only variants from My Cancer Genome.
Rights and permissions
About this article
Cite this article
Beaubier, N., Bontrager, M., Huether, R. et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat Biotechnol 37, 1351–1360 (2019). https://doi.org/10.1038/s41587-019-0259-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0259-z
This article is cited by
-
Multi-omic profiling of simultaneous ductal carcinoma in situ and invasive breast cancer
Breast Cancer Research and Treatment (2024)
-
An approach for improvement of the accuracy of cancer gene panel testing
International Journal of Clinical Oncology (2024)
-
Comparative genomic analysis of PIK3R1-mutated and wild-type breast cancers
Breast Cancer Research and Treatment (2024)
-
The Unseen Hand: AI-Based Prescribing Decision Support Tools and the Evaluation of Drug Safety and Effectiveness
Drug Safety (2024)
-
Sarcoma microenvironment cell states and ecosystems are associated with prognosis and predict response to immunotherapy
Nature Cancer (2024)