Abstract
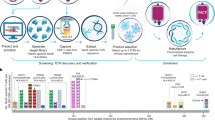
Targeting membrane proteins could improve the efficacy of T cell–based immunotherapies. To facilitate the identification of T cell targets, we developed a hybrid genetic screening system where the Sleeping Beauty (SB) transposon and single guide RNA cassette are nested in an adeno-associated virus (AAV). SB-mediated genomic integration of the single guide RNA cassette enables efficient gene editing in primary murine T cells as well as a screen readout. We performed in vivo AAV–SB-CRISPR screens for membrane protein targets in CD8+ T cells in mouse models of glioblastoma (GBM). We validated screen hits by demonstrating that adoptive transfer of CD8+ T cells with Pdia3, Mgat5, Emp1 or Lag3 gene editing enhances the survival of GBM-bearing mice in both syngeneic and T-cell receptor transgenic models. Transcriptome profiling, single cell sequencing, cytokine assays and T cell signaling analysis showed that Pdia3 editing in T cells enhances effector functions. Engineered PDIA3 mutant EGFRvIII chimeric antigen T cells are more potent in antigen-specific killing of human GBM cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data and statistics for non-NGS experiments such as tumor studies, flow cytometry, T7E1, qPCR, protein experiments and coculture assays are provided in an Excel table as Source Data. Genome sequencing data are deposited to the Sequence Read Archive with accession number PRJNA553676. Other data, reagents, methods, computational code and materials that support the findings of this research are available from the corresponding author upon reasonable request.
Code availability
Custom code used to support the findings of this research is available from the corresponding author upon reasonable request.
References
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568 (2014).
Kvistborg, P. et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8(+) T cell response. Sci. Transl. Med. 6, 254ra128 (2014).
Ribas, A. Tumor immunotherapy directed at PD-1. New Engl. J. Med. 366, 2517–2519 (2012).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Omuro, A. et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 20, 674–686 (2018).
O’Rourke, D. M. et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017).
Chow, R. D. et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 20, 1329–1341 (2017).
Wang, G. et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci. Adv. 4, eaao5508 (2018).
Mates, L. et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 (2009).
Colella, P., Ronzitti, G. & Mingozzi, F. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104 (2018).
Wang, D., Tai, P. W. L. & Gao, G. P. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Hogquist, K. A. et al. T-cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Workman, C. J., Dugger, K. J. & Vignali, D. A. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J. Immunol. 169, 5392–5395 (2002).
Workman, C. J. et al. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J. Immunol. 172, 5450–5455 (2004).
Anderson, A. C., Joller, N. & Kuchroo, V. K. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 44, 989–1004 (2016).
Huang, C. T. et al. Role of LAG-3 in regulatory T cells. Immunity 21, 503–513 (2004).
Smith, L. K. et al. Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312 e295 (2018).
Shibui, A. et al. Alteration of immune responses by N-acetylglucosaminyltransferase V during allergic airway inflammation. Allergol. Int. 60, 345–354 (2011).
Morgan, R. et al. N-acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J. Immunol. 173, 7200–7208 (2004).
Sugiura, K. & Stock, C. C. Studies in a tumor spectrum. II. The effect of 2,4,6-triethylenimino-s-triazine on the growth of a variety of mouse and rat tumors. Cancer 5, 979–991 (1952).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Schlager, C. et al. Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature 530, 349 (2016).
Khalil, D. N., Smith, E. L., Brentjens, R. J. & Wolchok, J. D. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 13, 273–290 (2016).
Ribas, A. et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol. 16, 908–918 (2015).
Preusser, M., Lim, M., Hafler, D. A., Reardon, D. A. & Sampson, J. H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat. Rev. Neurol. 11, 504–514 (2015).
Cloughesy, T. F. et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 25, 477–486 (2019).
Li, B. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17, 174 (2016).
Reardona, D. A. et al. Randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: Checkmate 143. Neuro Oncol. 19, 21–21 (2017).
Agarwalla, P., Barnard, Z., Fecci, P., Dranoff, G. & Curry, W. T. Sequential immunotherapy by vaccination with GM-CSF-expressing glioma cells and CTLA-4 blockade effectively treats established murine intracranial tumors. J. Immunother. 35, 385–389 (2012).
Fecci, P. E. et al. Systemic CTLA-4 blockade ameliorates glioma-induced changes to the CD4(+) T cell compartment without affecting regulatory T-cell function. Clin. Cancer Res. 13, 2158–2167 (2007).
Mathios, D. et al. Anti-PD-1 antitumor immunity is enhanced by local and abrogated by systemic chemotherapy in GBM. Sci. Transl. Med. 8, 370ra180 (2016).
Saha, D., Martuza, R. L. & Rabkin, S. D. Macrophage polarization contributes to glioblastoma eradication by combination immunovirotherapy and immune checkpoint blockade. Cancer Cell 32, 253–267.e5 (2017).
Kim, J. E. et al. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin. Cancer Res. 23, 124–136 (2017).
Dai, H., Wang, Y., Lu, X. & Han, W. Chimeric antigen receptors modified T-cells for cancer therapy. J. Natl Cancer Inst. 108, djv439 (2016).
Brown, C. E. et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N. Engl. J. Med. 375, 2561–2569 (2016).
Pellegatta, S. et al. Constitutive and TNFɑ-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: implications for CAR-T cell therapy. Sci. Transl. Med. 10, eaao2731 (2018).
Mount, C. W. et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M(+) diffuse midline gliomas. Nat. Med. 24, 572–579 (2018).
Bernards, R. Finding effective cancer therapies through loss of function genetic screens. Curr. Opin. Genet. Dev. 24, 23–29 (2014).
Okada, M. et al. Blockage of core fucosylation reduces cell-surface expression of PD-1 and promotes anti-tumor immune responses of T cells. Cell Rep. 20, 1017–1028 (2017).
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018).
Ting, P. Y. et al. Guide Swap enables genome-scale pooled CRISPR–Cas9 screening in human primary cells. Nat. Methods 15, 941–946 (2018).
Zhou, P. et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52–57 (2014).
Chen, R. et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338 (2014).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Demetriou, M., Granovsky, M., Quaggin, S. & Dennis, J. W. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 (2001).
Granovsky, M. et al. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat. Med. 6, 306–312 (2000).
Doench, J. G. et al. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Uren, A. G. et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 4, 789–798 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Pimentel, H., Bray, N. L., Puente, S., Melsted, P. & Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Acknowledgements
We thank C. Fuchs, D. Hafler, R. Montgomery, D. Rimm and M. Gunel for discussions. We thank S. Lam, L. Shen, Z. Bai, H. Ye, R. Kim and other members of the Chen laboratory, as well as various colleagues in the Department of Genetics, Systems Biology Institute, Cancer Systems Biology Center, MCGD Program, Immunobiology Program, BBS Program, Cancer Center and Stem Cell Center at Yale for assistance and/or discussion. We thank the Center for Genome Analysis, the Center for Molecular Discovery, Pathology Tissue Services, Histology Services, the High Performance Computing Center, West Campus Analytical Chemistry Core and West Campus Imaging Core, the Mass Cytometry Core and Keck Biotechnology Resource Laboratory at Yale for technical support. S.C. is supported by Yale SBI/Genetics Startup Fund, the NIH/NCI (grant nos. DP2CA238295, R01CA231112, U54CA209992-8697, R33CA225498, RF1048811, P50CA196530-A10805, P50CA121974-A08306), the Damon Runyon Dale Frey Award (grant no. DFS-13-15), the Melanoma Research Alliance (grant nos. 412806, 16-003524), St-Baldrick’s Foundation (grant no. 426685), the Breast Cancer Alliance, the Cancer Research Institute (CLIP), AACR (grant nos. 499395, 17-20-01-CHEN), The Mary Kay Foundation (grant no. 017-81), The V Foundation (grant no. V2017-022), the Ludwig Family Foundation, DoD (grant no. W81XWH-17-1-0235), the Sontag Foundation and the Chenevert Family Foundation. G.W. is supported by CRI Irvington and RJ Anderson Fellowships. X.D. is supported by a Revson Fellowship. M.B.D., R.D.C. and J.J.P. are supported by the Yale MSTP training grant from the NIH (grant no. T32GM007205).
Author information
Authors and Affiliations
Contributions
L.Y. designed the AAV–SB–CRISPR vector, developed the screening system and performed the majority of experiments in this study. J.J.P. analyzed the CRISPR screen and other high-throughput data. M.B.D. generated transgenic mice and optimized the screen. R.D.C. designed the Surf library. Q.Y., L.P., Y.D., J.G., G.W. and Y.E. assisted with various experiments. Y.D. and X.D. designed and generated the EGFRvIII CAR-T system. S.C. conceived the study, secured funding and supervised the work. L.Y., J.J.P. and S.C. prepared the manuscript with input from all of the other authors.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed related to the data in this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Splinkerette PCR identify genome integration of Sleeping Beauty transposon.
(a) Nextera indel analysis for Mll3 and B2m knock-out in mouse CD8+ T cells using AAV-sgRNA vectors. Representative mutations and their frequencies were shown around predicted sgRNA target sites. (b) Schematics of splinkerette PCR procedures. The steps include genomic DNA isolation, restriction enzyme digestion, adaptor ligation, PCR, NGS library prep, and sequencing. (c) Electrophoresis of the splinkerette PCR products. The gel within red dash line was gel purified for the Nextera library preparation and sequencing. (d) Representative SB transposon integration sites in the mouse genome. (e) A bar plot of splinkerette PCR read distribution for the number of integration sites along mouse chromosomes. (f) A bar plot showed splinkerette PCR read distribution for the number of integration sites according to functional annotation of genomic regions.
Supplementary Figure 2 Single cell RT-qPCR estimation of functional MOI of AAV-SB-CRISPR screen.
Single cell RT-qPCR detection of single T cell expressing functional sgRNAs, for the estimation of functional MOI with exact transduction parameters in the AAV-Surf screen. PBS treated single T cells were used as a negative control. Numbers of wells without T cells, with sgRNA- T cells and with sgRNA+ T cells were determined to estimate MOI.
Supplementary Figure 3 Penetrance estimation of intracranial brain tumor induction using GL261 and derivatives cell lines.
(a) Bar plot of mortality rate of mouse after intracranial GBM induction. Different GBM cell lines, GL261, GL261-Luc-Ova, and GL261-Luc, were used for brain injection, and each cell line injected with different cell number. (b) Bar plot of number of mice that met euthanasia endpoint due to brain tumor.
Supplementary Figure 4 AAV-Surf library transduced mouse CD8+ T cell surface phenotypes and TCR repertoires before and after adoptive transfer.
(a) Flow cytometry analysis of surface PD-1, Lag3, and Tim-3 expression after transduced with AAV-Vector and AAV-Surf virus. Result from one experiment. (b-c) Flow cytometry analysis of proportion of PD-1+ TILs. ns, non-significant. Data are shown as mean ± s.e.m., with individual data points. The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table. (d) A bar plot of T cell clonal composition from TCR sequencing. Pre injection, mouse CD8+ T cells transduced with AAV-Surf virus and cultured for 5 days; Post injection (TIL), AAV-Surf transduced T cells i.v. injected into GBM-bearing mice and isolated as TILs at day 6 after T cell injection. (e) A ring plot of TCR distribution for T cells before i.v. injection. Top TCR sequences were labeled in the plot. (f) A ring plot of TCR distribution for T cells after i.v. injection (TILs). Top TCR sequences were labeled in the plot.
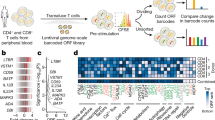
Supplementary Figure 5 AAV-CRISPR CD8+ T cell screens of membrane bound proteome knockouts in GBM.
(a) Schematics of hybrid AAV-SB-CRIPSR CD8+ T cell screen in a syngeneic mouse model of GBM, showing steps of naïve CD8+ T cell isolation, AAV library transduction, GBM cell transplantation, adoptive T cell transfer, brain and tumor isolation, and sgRNA readout by deep sequencing. (b) Representative in vivo imaging illustrating the growth of GL261-FLuc tumors in the brains of C57BL/6 mice. (c) Representative photos of mice after GBM transplantation and T cell treatment. Red circles indicated macrocephaly suggestive of growing brain tumors. (d) Survival plot of mice after GBM engraftment and adoptive transfer. C57BL/6J mice were transplanted with 1.2 x 106 GL261-FLuc into the lateral ventricle (LV). 4 x 106 OT-I;Cas9β CD8+ T cells were injected after 10 days of tumor engraftment. Survival significance was assessed by a log-rank Mantel-Cox test. The p-values and number of mice used in each group was indicated in the plots. The group of mice receiving adoptive transfer of T cells has significantly enhanced survival, where the effect of AAV-Surf is slightly stronger than Vector. DPI, days post tumor implantation. (e) Representative H&E stained brain sections from PBS, AAV-Vector and AAV-Surf groups. Areas within red dashed lines indicate brain tumors. Scale bar, 2 mm for whole brain sections, and 100 μm for zoom-in sections. These are representative images at the endpoint of survival thus not quantitative for comparison in terms of tumor burden. One experiment, n = 3 individual mice for PBS, n = 5 mice for vector, n = 11 mice for AAV-Surf. (f) Scatterplot of brain vs. cell sgRNA library representation of AAV-Surf shorter term screen experiment (max survival 20 days post injection). The most enriched sgRNAs in the brain are highlighted. Purple dash line, y = x curve; blue dash line, linear regression of the distribution of the 1,000 NTCs between the brain and cell samples. (g) Scatterplot of brain vs. cell sgRNA library representation of AAV-Surf longer term screen experiment (max survival 92 days post injection). The most enriched sgRNAs in the brain are highlighted. Purple dash line, y = x curve; blue dash line, linear regression of the distribution of the 1,000 NTCs between the brain and cell samples. The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
Supplementary Figure 6 Clonal GBM cell line generation and representative mouse GBM histology with single gene knockout adoptive transfer.
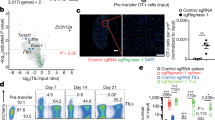
(a) Flow cytometry analysis of GL261-FLuc-mCh-cOVA clones for cOVA expression level. Two independent experiments. (b) T7EI assay indicated gene editing of Mgat5 and Pdia3 with AAV-SB100x vector. Multiple, at least five independent experiments. (c) T7EI assay for mouse T cells treated with sgPdia3 on top 4 predicted off-target sites. Two experiments. (d) RT-qPCR of Mgat5 and Pdia3 after infected with AAV6 carrying specific gene-targeting sgRNAs in AAV-SB100x vector. Unpaired t test was used for the significance assessment, AAV-Vector vs. AAV-sgMgat5, p = 0.0242. * p < 0.05. (e) Schematic of the therapeutic efficacy testing strategy for top candidates identified from the AAV-Surf screens, using adoptive transfer of single gene edited CD8+ T cells in a syngeneic mouse model of GBM. (f) Survival plots of the top candidate validations in a syngeneic mouse model of GBM. C57BL/6J mice were engrafted with 2 x 105 GL261 cancer cells, and adoptive transfer treatment was performed after 10 days of tumor engraftment by intravenous injection of 6 x 105 Cas9β CD8+ T cells infected with AAV-Vector, AAV-sgLag3, AAV-sgMgat5, and AAV-sgPdia3. Survival significance was assessed by a log-rank Mantel-Cox test. (g) Representative H&E stained brain sections from AAV-Vector and AAV-sgRNA single knockout groups in C57BL/6J mice. Scale bar, 100 μm. These are representative images at the endpoint of survival thus not quantitative for comparison in terms of tumor burden. One experiment, n = 8 individual mice for each group. (h) Representative H&E stained brain sections from AAV-Vector and AAV-sgRNA single knockout groups in Rag1-/- mice. Scale bar, 100 μm. These are representative images at the endpoint of survival thus not quantitative for comparison in terms of tumor burden. One experiment, n = 8 mice for AAV-Vector, n = 10 mice for AAV-sgMgat5, n = 9 for AAV-sgPdia3, and n = 8 for AAV-sgEmp1. The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
Supplementary Figure 7 Representative luciferase imaging for tumor burden quantification.
(a) Quantification of tumor burden as total luciferase flux at days 12, 16, and 18. Day 20 and 22 data points were not used for statistics because most mice in the AAV-Vector group had already reached endpoint. Mice being imaged, n = 8 for Vector, n = 8 for sgMgat5, n = 5 for sgPdia3. Data are shown as mean ± s.e.m. plus individual data points on bar graphs. (b) Quantification of tumor burden as total luciferase flux at days 14, 15, 17, 19, and 22 after tumor induction. Mice being imaged, n = 7 for Vector, n = 7 for sgMgat5, n = 8 for sgPdia3, n = 8 for sgMgat5+sgPdia3. Data are shown as mean ± s.e.m. plus individual data points on bar graphs. The numbers of mice that had reached endpoint, euthanized, and therefore removed from the imaging group are indicated on the top table.
Supplementary Figure 8 Single cell RNA-seq profiling of Pdia3 KO CD8+ T cells.
(a) t-SNE plot of hierarchical clustering of scRNA-seq results. (b) Volcano plot of scRNA-seq of mouse CD8+ T cells after Pdia3 knockout. Pdia3 is a significantly downregulated gene after infection with AAV-sgPdia3. A total 9,193 single cells were captured and their transcriptomes were sequenced for the AAV-sgPdia3 (n = 4,549 single cells) and AAV-Vector (n = 4,644 single cells) treated CD8 T cells. Two-sided Wilcoxon signed-rank test by gene, with p-values adjusted by Benjamini & Hochberg.
Supplementary Figure 9 Immune marker analysis of PDIA3 KO human CD8+ T cells by CyTOF.
(a) t-SNE plot clustered by samples, showing that the 3 PDIA3 KO samples grouped with each other, the 3 WT samples also grouped with each other, and that PDIA3 KO samples and WT samples formed distinct groups. (b) t-SNE plot of CyTOF data with k-means clustering revealing 10 major clusters. (c) t-SNE and violin plots of CyTOF data of CD127 / IL7R, FAS / CD95, 4-1BB / CD137 and TIM-3 / HAVCR2 at the surface protein level, for both PDIA3 KO and wildtype single human CD8+ T cells (n = 3 replicates each, sampled 7,000 cells per replicate for comparison). Violins show kernel probability density on side, and boxplot is standard, i.e. middle band is median, hinges/ends of box are interquartile range (25% and 75% quantiles), lower whisker = smallest observation greater than or equal to lower hinge - 1.5 * IQR, upper whisker = largest observation less than or equal to upper hinge + 1.5 * IQR. Wilcoxon test, two-sided, p value adjusted by Benjamini & Hochberg method. KO vs WT: p = 1.91e-81 for CD127 / IL7R, p = 0.1147 for FAS / CD95, p = 6.75e-83 for 4-1BB / CD137, and p = 0 for TIM-3 / HAVCR2.
Supplementary Figure 10 TIDE analysis of PDIA3-related T cell dysfunction in human cancer patients.
(a–c) Analyses of PDIA3 expression signatures linked to in cytotoxic T lymphocyte (CTL) – associated survival benefits in patients, where high-level of PDIA3 abolishes or weakens the overall survival benefit of CTL-high patients with GBM (a), TNBC (b), or LUAD (c). X-axis is overall survival in months. Y-axis is survival fraction. Specific statistical tests using TIDE (Methods). (d) Analyses of PDIA3 expression signatures linked to survival benefits of patients treated with an immune-checkpoint antibody (Ipilimumab, anti-CTLA4), in melanoma. X-axis is overall survival in days. Y-axis is survival fraction. Specific statistical tests were performed using TIDE (Methods).
Supplementary Figure 11 Representative flow cytometry gating.
(a) Gating of mouse Ifnγ+ CD8+ T cells. (b) Gating of human IFNγ+ CD8+ T cells.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Methods.
Supplementary Datasets
Supplementary Datasets 1–9.
Supplementary Tables
Supplementary Tables 1–11.
Source Data for Figs. 1–6 and Supplementary Figs. 4–7
Source Data for Figs. 1–6 and Supplementary Figs. 4–7
Rights and permissions
About this article
Cite this article
Ye, L., Park, J.J., Dong, M.B. et al. In vivo CRISPR screening in CD8 T cells with AAV–Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat Biotechnol 37, 1302–1313 (2019). https://doi.org/10.1038/s41587-019-0246-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0246-4
This article is cited by
-
Functional CRISPR screens in T cells reveal new opportunities for cancer immunotherapies
Molecular Cancer (2024)
-
CRISPR–Cas9 applications in T cells and adoptive T cell therapies
Cellular & Molecular Biology Letters (2024)
-
Protein disulfide-isomerase A4 confers glioblastoma angiogenesis promotion capacity and resistance to anti-angiogenic therapy
Journal of Experimental & Clinical Cancer Research (2023)
-
CRISPR/Cas9 system: recent applications in immuno-oncology and cancer immunotherapy
Experimental Hematology & Oncology (2023)
-
Targeting lymph node delivery with nanovaccines for cancer immunotherapy: recent advances and future directions
Journal of Nanobiotechnology (2023)