Abstract
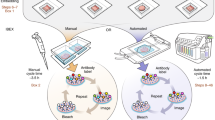
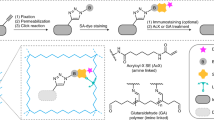
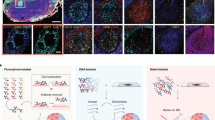
Spatial mapping of proteins in tissues is hindered by limitations in multiplexing, sensitivity and throughput. Here we report immunostaining with signal amplification by exchange reaction (Immuno-SABER), which achieves highly multiplexed signal amplification via DNA-barcoded antibodies and orthogonal DNA concatemers generated by primer exchange reaction (PER). SABER offers independently programmable signal amplification without in situ enzymatic reactions, and intrinsic scalability to rapidly amplify and visualize a large number of targets when combined with fast exchange cycles of fluorescent imager strands. We demonstrate 5- to 180-fold signal amplification in diverse samples (cultured cells, cryosections, formalin-fixed paraffin-embedded sections and whole-mount tissues), as well as simultaneous signal amplification for ten different proteins using standard equipment and workflows. We also combined SABER with expansion microscopy to enable rapid, multiplexed super-resolution tissue imaging. Immuno-SABER presents an effective and accessible platform for multiplexed and amplified imaging of proteins with high sensitivity and throughput.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study will be provided by the corresponding authors upon reasonable request.
Code availability
The deep learning algorithm and test dataset for automated identification of nuclear contours in tonsil tissues are available at https://github.com/HMS-IDAC/UNet. The MATLAB code for nuclear segmentation is available at https://github.com/HMS-IDAC/SABERProbMapSegmentation.
References
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Levenson, R. M., Borowsky, A. D. & Angelo, M. Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab. Invest. 95, 397–405 (2015).
Giesen, C. W. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–425 (2014).
Wei, L. et al. Super-multiplex vibrational imaging. Nature 544, 465–470 (2017).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042 (2018).
Wang, Y. et al. Rapid sequential in situ multiplexing with DNA exchange imaging in neuronal cells and tissues. Nano Lett. 17, 6131–6139 (2017).
Schueder, F. et al. Universal super-resolution multiplexing by DNA exchange. Angew. Chem. Int. Ed. Engl. 56, 4052–4055 (2017).
Jungmann, R. et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318 (2014).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Lundberg, E. et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450 (2010).
Bobrow, M., Litt, G. J., Shaughnessy, K. J., Mayer, P. C. & Conlon, J. The use of catalyzed reporter deposition as a means of signal amplification in a variety of formats. J. Immunol. Methods 150, 145–149 (1992).
Yarilin, D. et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Sci. Rep. 5, 9534 (2015).
Stack, E. C., Foukas, P. G. & Lee, P. P. Multiplexed tissue biomarker imaging. J. Immunother. Cancer 4, 9 (2016).
Schweitzer, B. et al. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl Acad. Sci. USA 97, 10113–10119 (2000).
Nagendran, M., Riordan, D. P., Harbury, P. B. & Desai, T. J. Automated cell-type classification in intact tissues by single-cell molecular profiling. eLife 7, e30510 (2018).
Deng, R. et al. DNA-sequence-encoded rolling circle amplicon for single-cell RNA imaging. Chem 4, 1373–1386 (2018).
Chen, Y. et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217, 4025–4048 (2018).
Pachl, C. et al. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. J. Acquir. Immune Defic. Syndr. 8, 446 (1995).
Kern, D. et al. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 34, 3196–3202 (1996).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Dirks, R. M. & Pierce, N. A. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. USA 101, 15275–15278 (2004).
Choi, H. M. et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212 (2010).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018).
Wang, Y., Xie, W., Kohman, R. E. & Church, G. M. Multiplexed imaging using same species primary antibodies with signal amplification. Preprint at https://doi.org/10.1101/274456 (2018).
Lin, R. et al. A hybridization-chain-reaction-based method for amplifying immunosignals. Nat. Methods 15, 275–278 (2018).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2018).
Kishi, J. Y. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 16, 533–544 (2019).
Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science 347, 543–548 (2015).
Nave, H., Gebert, A. & Pabst, R. Morphology and immunology of the human palatine tonsil. Anat. Embryol. 204, 367–373 (2001).
Slijkerman, R. W. et al. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog. Retin. Eye Res. 48, 137–159 (2015).
Speel, E. J., Hopman, A. H. & Komminoth, P. Amplification methods to increase the sensitivity of in situ hybridization: play card(s). J. Histochem. Cytochem. 47, 281–288 (1999).
Clutter, M. R., Heffner, G. C., Krutzik, P. O., Sachen, K. L. & Nolan, G. P. Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry A 77, 1020–1031 (2010).
Zadeh, J. N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Uhlen, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Chang, J. B. et al. Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017).
Truckenbrodt, S. et al. X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19, e45836 (2018).
Dani, A., Huang, B., Bergan, J., Dulac, C. & Zhuang, X. Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 (2010).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Snyder, M. M. et al. Mapping the human body at cellular resolution—the NIH Common Fund Human BioMolecular Atlas program. Preprint at https://arxiv.org/abs/1903.07231 (2019).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Takada, S. E. & Engleman, E. G. Evidence for an association between CD8 molecules and the T cell receptor complex on cytotoxic T cells. J. Immunol. 139, 3231–3235 (1987).
Beck, M. et al. The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011).
Bernstein, H. G. et al. Regional and cellular distribution of neural visinin-like protein immunoreactivities (VILIP-1 and VILIP-3) in human brain. J. Neurocytol. 28, 655–662 (1999).
Haverkamp, S. & Wassle, H. Immunocytochemical analysis of the mouse retina. J. Comp. Neurol. 424, 1–23 (2000).
Pierce, N. A. & Dirks, R. M. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 24, 1664–1677 2003).
Dirks, R. M. & Pierce, N. A. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 25, 1295–1304 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Allan, C. et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253 (2012).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. Preprint at https://arxiv.org/abs/1505.04597 (2015).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Preprint at https://arxiv.org/abs/1502.03167 (2015).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. Preprint at https://arxiv.org/abs/1512.03385 (2015).
Acknowledgements
We thank C. Cepko, P. Sorger, Z. Maliga and J. Lin for discussion. We thank the Neurobiology Department and the Neurobiology Imaging Facility for instrument support. This facility is supported in part by the Neural Imaging Center as part of NINDS P30 Core Center grant NS072030. We thank M. Manesse, T. Archivald and D. Bowman for help with the FFPE samples, and I. Goldaracena for comments on the manuscript. We thank S. Wang for providing neuronal cultures. This work was supported by grants from the National Institutes of Health (under grants NIH Common Fund 1UG3HL145600/HuBMAP, 1R01EB018659, 1U01MH106011, 1DP1GM133052 and 1R01GM124401), the Office of Naval Research (under grants N00014-16-1-2410, N00014-16-1-2182 and N00014-18-1-2549), the National Science Foundation (under grant CCF-1317291), Harvard Medical School Dean′s Initiative and Wyss Institute′s Molecular Robotics Initiative to P.Y. G.M.C. was supported by NIH grants (R01NS083898, RM1 HG008525 and R01MH113279), and P.S.K. was supported by an NIH grant (1R01MH113349). J.Y.K. was supported by a National Science Foundation Graduate Research Fellowship. B.J.B. was supported by a Damon Runyon Cancer Research Foundation Fellowship. S.W.L. was supported by HHMI and the National Institutes of Health (grant 5K99EY028215-02). W.X., Y.Z. and S.Y. were partially supported by visiting undergraduate student funding from Fudan University, Tsinghua University and Shanghai Jiaotong University, respectively. S.K.S. was supported by long-term postdoctoral fellowships from EMBO (ALTF 1278- 2015) and the Human Frontier Science Program (HFSP) (LT000048/2016-L).
Author information
Authors and Affiliations
Contributions
S.K.S. and Y.W. conceived the study, performed experiments, analyzed the data and wrote the manuscript. J.Y.K. contributed to conceptualization of the study, protocol optimization and experimental design. A.Z., Y.Z., W.X., S.Y. and M.L. provided experimental assistance. K.K., C.Y. and M.C. performed data analysis and visualization. B.J.B. contributed to protocol optimization. S.W.L. provided the retina samples for staining. G.P. contributed to contextualization of SABER application, and provided guidance and tissues for the FFPE study. E.S.B. contributed to the expansion study. P.S.K. contributed to the neuronal study. G.M.C. provided scientific guidance and contributed to study supervision. P.Y. conceived and supervised the study, and wrote the manuscript. All authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.K.S., Y.W., J.Y.K., B.J.B. and P.Y. are inventors for a US patent application based on this work (PCT/US2018/013019). P.Y. is a co-founder of Ultivue, Inc. and NuProbe Global.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Optional purification of DNA-conjugated antibodies using a toehold displacement-mediated DNA affinity pull-down.
(a) Schematic of the pull down assay. Biotin-modified capture DNA strands (biotin-c-b*, where c is 15-nucleotides and b* is 16-nucleotides) are attached to streptavidin beads, and used to pull down DNA-conjugated antibodies after antibody-bridge DNA conjugation (bridge strand sequence: a-b, where b is 16 nucleotides). The attached antibodies are dissociated from the beads using toehold displacement strands (c*-b) that compete with the capture strands (Zhang and Winfree, 2009) on antibodies. While optional for Immuno-SABER, we found that purifying the DNA-conjugated antibodies via pull-down and toehold-mediated displacement may be helpful to improve the signal for select antibodies. (b) Visualization of purification products using an SDS-PAGE gel assay. After DNA conjugation, the majority of antibodies were conjugated with 0, 1 or 2 DNA oligos per antibody. After purification, antibodies without DNA were removed. (c) Plot of protein densities for the bands in b. The band corresponding to the removed unconjugated antibodies is marked with an arrow. Ref: Zhang, D.Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc 131, 17303-17317 (2009).
Supplementary Figure 2 Resolution and penetration controls for Immuno-SABER.
(a) ~2.5 µm long lineplots (red) were made over the thin tubules to estimate the observed resolution. Yellow-boxed background regions were used to estimate the background for subtraction. (b) A typical line plot along a tubule and the Gaussian fit, where full-width-half-maximum (FWHM) was calculated as 371 nm. (c) Mean FWHM values were calculated for 30-45 lineplots from cells stained with Immuno-SABER or fluorophore-conjugated secondary antibodies samples and the distribution was displayed as a box plot. A similar calculation was performed for 200 nm fluorescent beads. Mean FWHM was not significantly different (p value is 0.360 for Immuno-SABER and 0.335 for conventional secondary antibody staining, two-sample t-test comparison to the bead sample). Box-plots are drawn with center line, median; box limits, upper and lower quartiles; whiskers, min and max values capped at 1.5× interquartile range. (d) Visualization of Collagen IV and Vimentin at multiple depths of the whole mount mouse retina shown in Fig. 2f. Selected confocal planes are shown. Vimentin stains the Muller cells and Collagen IV stains the blood vessels, both of which are localized predominantly in the segments from nerve fiber layer to outer plexiform layer (~100 µm) of the retina (Slijkerman et al., 2015). Hence it should be noted that, although the entire whole-mount retina is about ~180 μm, the target signal comes from roughly one half of the retina section, from the nerve fiber layer to the outer plexiform layer. Ref: Slijkerman, R.W. et al. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog Retin Eye Res 48, 137-159 (2015).
Supplementary Figure 3 Higher signal amplification via branching.
(a) Primary PER concatemers can be targeted by secondary concatemers to form a branched structure which amplifies the signal further by presenting additional binding sites for the imagers. (b-c) Representative images for linear and branched SABER amplification are shown for different preparations: (b) CD8a staining in human tonsil FFPE sections (single plane large area scans with 20× objective, cropped to show a CD8a-rich interfollicular zone region, (c) Cone arrestin staining in mouse retina cryosections (max projections from confocal z stacks). (d) Level of signal amplification by branched Immuno-SABER over linear was quantified by measuring the background-subtracted mean fluorescence for several regions of interest in the tissues and expressed as fold amplification over linear SABER. For CD8a FFPEs, n = 144 (for linear) and 84 (for branched) rectangular ROIs (each covering 0.03-1.20 mm2 tissue regions; consecutive sections are used for the two conditions). For cone arrestin, n = 6 images from 2 retina samples. Error bars, s.e.m. (e) Alpha-tubulin staining (Alexa647) in cultured BS-C-1 cells (max projections from confocal z stack). (f) Mean FWHM values were calculated for 43 lineplots from cells stained with branched Immuno-SABER. For comparison values for conventional staining is also included. Box-plots are drawn with center line, median; box limits, upper and lower quartiles; whiskers, min and max values capped at 1.5× interquartile range. Mean FWHM for branching was not significantly different than the sub-diffraction 200 nm bead samples (plotted in Supplementary Fig. 2c) (p value is 0.303, two-sample t-test comparison to the bead sample).
Supplementary Figure 4 Accessibility, concatemer length, and quantitativeness controls for Immuno-SABER.
(a) Experiment design: HeLa cell preparations were stained with anti-Lamin B antibodies and oligo-conjugated secondary antibodies. Concatemers of different sizes (short: 350 nt, medium: 450 nt, long: 700 nt; length estimations are based on the gel run with respect to dsDNA ladder) were then hybridized to the antibodies. For this case, concatemers that contain an extra orthogonal binding site (for i.27*, as baseline imager) before the primer domain (p.28) were utilized. Samples were imaged first with only the baseline imager, then with the amplifier imager (i.28*). Imaging was done at 100× and 22-25 plane z stacks were acquired by an epifluorescence microscope. (b) Nuclei were segmented based on DAPI and mean Lamin B fluorescence intensity per nuclear pixel was calculated. n = 23-45 cells (individual sample sizes are written in parenthesis above each bar). Error bars, s.e.m. (c) For reference, the same target was also stained with commercial fluorophore-conjugated secondaries that on average bear 5× Alexa647 fluorophores and the fluorescence was measured the same way. (d) Representative images show the maximum projections for DAPI and Lamin B staining (Alexa647) for baseline and amplification conditions. The intensity scaling for each row is note on the left-hand side. (e) Secondary antibody-fluorophore images are shown for comparison. (f) Branching experiment design: A separate set of cells were similarly imaged with the baseline imager after hybridization of short (350 nt) or long primary concatemers (700 nt). This was followed by hybridization of secondary concatemers (short: 250 nt, long: 450 nt). Secondary concatemers also contained an orthogonal binding site (for i.30*) before the primer domain (p.25*). Baseline for branching (post-linear) was imaged by i.30*, followed by branched amplification with i.25*. (g) Lamin B fluorescence intensity per nuclear pixel was calculated as in b and is shown with the bar plot (left axis) overlaid with the scatter plot showing the distribution in the dataset. The dot plot above (magenta, right axis) shows the coefficient of variation for all the conditions. Note that the imagers bind dimers of primer units, whereas branches bind trimers for higher stability during exchange rounds. Therefore compared to the i.28* staining in previous panels, i.30* is expected to yield 1.5-fold lower signal (blue bars in b and g). (h) Representative images show the maximum projections corresponding to the conditions in g. All images are acquired under comparable conditions, and are displayed at the given intensity scaling for each amplification level.
Supplementary Figure 5 Additional images for branching.
(a) Machine-learning based nucleus segmentation for signal quantification: The deep learning model was trained with a manually annotated dataset to enable automatic identification of nuclear contours to be followed by watershed segmentation. The image highlights nuclear contours (right side of the image) from DAPI staining (left side). Right panel: Watershed segmentation was used to segment (pink) the pixels corresponding to nuclei of each cell. (b-c) Images display a typical germinal center in human FFPE tonsil samples stained for Ki-67 (Alexa647, red) by Immuno-SABER. DAPI stain (blue) is shown for reference. Qualitatively similar amplification levels were obtained by long and short hybridization times (at 37 °C) for the primary concatemer and branching concatemer (75 min each). (d) Iterative SABER of SV2 (Alexa647) in a 40 μm mouse retina cryosection. (e) For comparison SV2 staining with TSA was performed using mono HRP conjugated secondary antibodies. (f) Zoom-out and zoom-in views of the high-magnification confocal images in Fig. 3f displayed at different scaling ranges for comparative visualization. 10 min TSA amplification is included for further comparison. (g) Application of tyramide-Alexa647 for the maximum recommended incubation of 10 min. The germinal center image on the left is scaled in the same range with Fig. 3d. Zoom-in on the right is included to display the significant blurring of the signal at 10 min incubation.
Supplementary Figure 6 Sequence validation for Exchange-SABER.
(a) 32 SABER sequences were extended to ~650 bases in vitro and examined by gel shift assay visualized with SybrGold on 6% denaturing PAGE gels. Qualitatively, 18 of the 32 sequences (such as #29) displayed a broader distribution with a ladder of shorter products visible albeit these bands being much dimmer than the predominant concatemer band. Although being extended, one sequence (#51) had a more even distribution in the upper length regime without a clear predominant band. Based on these distributions, particular applications may favor a subset of the sequence library, i.e. high efficiency primers (for example primers 27, 28, 30, 31, 37, 38, 41, 44, 47, 49, 50, 54) may be more favorably utilized for more quantitative experiments or for lower abundance proteins. (b-d) In situ performance and crosstalk analysis. BS-C-1 cells were stained with bridge DNA-conjugated antibodies targeting α-Tubulin on a 96-well plate. Concatemers extended from each primer were hybridized to the bridges creating an array of wells labeled with primer sequences p.25-p.56. For each concatemer two sets of wells were prepared: (i) cognate group to be incubated with the corresponding imager strands, and (ii) crosstalk group where we added mixtures of imagers except the cognate imager strand. For each primer (e.g. p.25), both cognate and crosstalk wells were prepared by either applying the corresponding Alexa647-imager (e.g. i.25*) or all the imagers except the cognate one (e.g. -i.26* to i.56*). Images were captured in 16-bit (0-65,535). Representative images are shown for Primer 25 (p.25) (c) and Primer 27 (p.27) concatemers (d). Crosstalk images are displayed with two different intensity scales to render the crosstalk signal visible. The fluorescence signals were quantified and plotted in the log scale and displayed as a heatmap. Consistent with the in vitro gel shift assay in panel a, sequences that had lower extension efficiency (particularly primers 29, 32, and 51) tended to yield less fluorescence signal compared to sequences that that extend with higher efficiency. Non-negligible crosstalk signal was only detected for Primer 27 concatemer (red box). (e) Crosstalk analysis of primer sequence p.27. BS-C-1 cells were fixed and stained with DNA-conjugated antibodies targeting alpha-Tubulin. Concatemers extended from primer sequence p.27 were hybridized to the antibodies, followed by addition of non-cognate imager strands. We first grouped every five imager strands and determined that p.27 had crosstalk with imager 44-49. We then tested individual imager strands from imager 44-49 and determined the strand responsible for crosstalk as imager strand 48 (i.48*), which is excluded from the library for further multiplexed imaging in presence of p.27.
Supplementary Figure 7 Controls for exchange imaging of FFPE human tonsil sections.
(a) Imagers can efficiently removed within 10 min by washing the sample with 50% formamide in PBS, as shown in before and after wash images of the same section stained for Ki-67 (red) with linear amplification, imaged and displayed under the same conditions. DAPI stain is shown in blue. (b) Under this wash condition (10 min with 50% formamide in PBS at RT) the concatemers are not displaced, as shown by exchange imaging of CD8a by re-binding the imagers to the same target and re-imaging the tissue section (linear amplification).
Supplementary Figure 8 Control experiments for highly multiplexed imaging in mouse retina sections.
(a) Comparison of antibody staining patterns before and after DNA conjugation. The images for unconjugated antibodies were taken using conventional fluorophore-conjugated secondary antibody labeling, and the images for antibodies after conjugation were taken using primary antibody-Immuno SABER labeling (images are displayed at individual contrast levels). (b) Efficiency of washing to remove the imager strands. A 30 μm mouse retina section was stained with DNA-conjugated SV2 antibodies and imaged using Alexa647-i.26* imager with a widefield microscope. Imager strands were washed using 0.1× PBS with 30% formamide at room temperature for 3×10 min. Before and after images were taken using the same imaging setting. The fluorescence intensity of indicated yellow line was measured using FIJI. (c) Washing conditions maintained sample integrity without signal loss. SV2 in mouse retina sections was imaged for 3 rounds and the correlation coefficient was calculated. The correlation coefficient between the images was above 0.95, suggesting the washing condition is sufficiently mild and non-disruptive. (d-e) Validation of the VLP1 and calretinin antibodies with conventional indirect immunostaining: The VLP1 (EnCor-mouse) and calretinin (EnCor-mouse) that were used for the multiplexing experiments (marked with asterisk) in Fig. 5 were tested for specificity by co-staining with other antibodies targeting the same targets, followed by visualization using fluorophore-conjugated secondary antibodies. (f) Three cell subtypes (marked with arrows, I: VLP1+ and Calretinin+, II: VLP1- and Calretinin+, III: VLP1+ and Calretinin–) identified in the multiplexed mouse retina imaging experiment were verified using conventional immunostaining with unconjugated primary antibodies (VLP1 by EnCor-rabbit and Calretinin by EnCor-mouse) and fluorophore-conjugated secondary antibodies.
Supplementary Figure 9 Other sample orientations and alternative reduction-mediated fluorophore removal timeline for Expansion-SABER.
(a) Additional post-expansion images of neuronal synapses showing different orientations, as in Fig. 6b. (b) High-zoom comparison of Vimentin and Calretinin signals with and without expansion. The image with expansion is the overlay of the Vimentin and Calretinin images from Fig. 6c, whereas the image without expansion was derived from images in Fig. 5a. Scale bar indicates the physical size after expansion (~3-fold). (c) Alternative fluorophore removal strategy using TCEP reduction in thick expanded samples using TCEP. A mouse retina section stained with SV2 was expanded and visualized with disulfide bond modified imager strands. The fluorophores (Alexa 647) on the imager strands were cleaved using TCEP reduction. The fluorescence signal was monitored using a confocal microscope in a time course for 10 min. (d) Quantification of fluorescence signals in c before and after TCEP reduction.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Supplementary Tables 1–5 and Supplementary Notes 1–3
Supplementary Protocols
Step-by-step instructions for Immuno-SABER
Rights and permissions
About this article
Cite this article
Saka, S.K., Wang, Y., Kishi, J.Y. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol 37, 1080–1090 (2019). https://doi.org/10.1038/s41587-019-0207-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0207-y
This article is cited by
-
Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases
Genome Medicine (2024)
-
Highly sensitive SERS platform for pathogen analysis by cyclic DNA nanostructure@AuNP tags and cascade primer exchange reaction
Journal of Nanobiotechnology (2024)
-
Thermal-plex: fluidic-free, rapid sequential multiplexed imaging with DNA-encoded thermal channels
Nature Methods (2024)
-
Multiplex protein imaging in tumour biology
Nature Reviews Cancer (2024)
-
Ultra high content analyses of circulating and tumor associated hybrid cells reveal phenotypic heterogeneity
Scientific Reports (2024)