Abstract
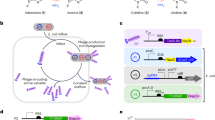
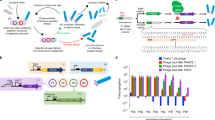
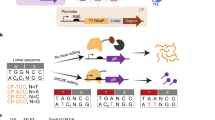
Base editors use DNA-modifying enzymes targeted with a catalytically impaired CRISPR protein to precisely install point mutations. Here, we develop phage-assisted continuous evolution of base editors (BE–PACE) to improve their editing efficiency and target sequence compatibility. We used BE–PACE to evolve cytosine base editors (CBEs) that overcome target sequence context constraints of canonical CBEs. One evolved CBE, evoAPOBEC1-BE4max, is up to 26-fold more efficient at editing cytosine in the GC context, a disfavored context for wild-type APOBEC1 deaminase, while maintaining efficient editing in all other sequence contexts tested. Another evolved deaminase, evoFERNY, is 29% smaller than APOBEC1 and edits efficiently in all tested sequence contexts. We also evolved a CBE based on CDA1 deaminase with much higher editing efficiency at difficult target sites. Finally, we used data from evolved CBEs to illuminate the relationship between deaminase activity, base editing efficiency, editing window width and byproduct formation. These findings establish a system for rapid evolution of base editors and inform their use and improvement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Key plasmids from this work will be available from Addgene (depositor, D.R.L.) and other plasmids are available upon request. All unmodified reads for sequencing-based data in the manuscript are available from the NCBI Sequence Read Archive, accession number PRJNA511456. Figures 4b, 5 and 6, Supplementary Table 3 and Supplementary Figs. 8–14, 16 and 18–22 are based on processing of sequencing data. Protein sequences used for Supplementary Fig. 17 are supplied as Supplementary Data 1.
Change history
12 August 2019
A Correction to this paper has been published: https://doi.org/10.1038/s41587-019-0253-5
References
Cornu, T. I., Mussolino, C. & Cathomen, T. Refining strategies to translate genome editing to the clinic. Nat. Med. 23, 415–423 (2017).
Webber, B.R. et al. Multiplex human T cell engineering without double-strand break induction using the Cas9 base editor system. Blood 132, 3495 (2018).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotech. 36, 843–846 (2018).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729–aaf8729 (2016).
Zafra, M. P. et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotech. 36, 888 (2018).
Gehrke, J. M. et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotech. 36, 977–982 (2018).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotech. 35, 371–376 (2017).
Najm, F. J. et al. Orthologous CRISPR–Cas9 enzymes for combinatorial genetic screens. Nat. Biotech. 36, 179–189 (2018).
Badran, A. H. & Liu, D. R. In vivo continuous directed evolution. Curr. Opin. Chem. Biol. 24, 1–10 (2015).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Badran, A. H. et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533, 58–63 (2016).
Bryson, D. I. et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Bio. 13, 1253–1260 (2017).
Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Bio. 10, 216–222 (2014).
Dickinson, B. C., Leconte, A. M., Allen, B., Esvelt, K. M. & Liu, D. R. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc. Natl Acad. Sci. USA 110, 9007–9012 (2013).
Dickinson, B. C., Packer, M. S., Badran, A. H. & Liu, D. R. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat. Commun. 5, 5352 (2014).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Hubbard, B. P. et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Chem. Bio. 12, 939–942 (2015).
Leconte, A. M. et al. A population-based experimental model for protein evolution: effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry 52, 1490–1499 (2013).
Packer, M. S., Rees, H. A. & Liu, D. R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 8, 956 (2017).
Wang, T., Badran, A. H., Huang, T. P. & Liu, D. R. Continuous directed evolution of proteins with improved soluble expression. Nat. Chem. Biol. 14, 972–980 (2018).
Roth, T., Woolston, B., Stephanopoulos, G. & Liu, D. R. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth. Biol. 8, 796–806 (2019).
Raindlová, V. et al. Influence of major-groove chemical modifications of DNA on transcription by bacterial RNA polymerases. Nucleic Acids Res. 44, 3000–3012 (2016).
Karzai, A. W., Roche, E. D. & Sauer, R. T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7, 449–455 (2000).
Lykke-Andersen, J. & Christiansen, J. The C-terminal carboxy group of T7 RNA polymerase ensures efficient magnesium ion-dependent catalysis. Nucleic Acids Res. 26, 5630–5635 (1998).
Rakonjac, J., Bennett, N. J., Spagnuolo, J., Gagic, D. & Russel, M. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Iss. Mol. Biol. 13, 51–76 (2011).
Zinder, N. D. & Boeke, J. D. The filamentous phage (Ff) as vectors for recombinant DNA–a review. Gene 19, 1–10 (1982).
Iwai, H., Züger, S., Jin, J. & Tam, P.-H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 (2006).
Beale, R. C. L. et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337, 585–596 (2004).
Navaratnam, N. et al. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol. 275, 695–714 (1998).
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 41, 578–594 (2016).
Kohli, R. M. et al. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284, 22898–22904 (2009).
Lada, A. G. et al. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry 76, 131–146 (2011).
St Martin, A. et al. A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC–Cas9 or cleavage by Cas9 in living cells. Nucleic Acids Res. 9, 229–210 (2018).
Wang, X. et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotech. 36, 946–949 (2018).
Manji, S. S. M., Miller, K. A., Williams, L. H. & Dahl, H.-H. M. Identification of three novel hearing loss mouse strains with mutations in the Tmc1 gene. The Am. J. Pathol. 180, 1560–1569 (2012).
Liu, C.-C., Liu, C.-C., Kanekiyo, T., Xu, H. & Bu, G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature Rev. Neurol. 9, 106–118 (2013).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 337, eaas9129–eaas9128 (2018).
Rigoli, L., Bramanti, P., Di Bella, C. & De Luca, F. Genetic and clinical aspects of Wolfram syndrome 1, a severe neurodegenerative disease. Pediatric Res. 83, 921–929 (2018).
Hardy, C. et al. Clinical and molecular genetic analysis of 19 Wolfram syndrome kindreds demonstrating a wide spectrum of mutations in WFS1. Am. J. Hum. Genet. 65, 1279–1290 (1999).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotech. 33, 187–197 (2014).
Scheben, A. & Edwards, D. Towards a more predictable plant breeding pipeline with CRISPR/Cas-induced allelic series to optimize quantitative and qualitative traits. Curr. Opin. Plant Biol. 45, 218–225 (2018).
Urnov, F. D., Ronald, P. C. & biotechnology, D. C. N. A call for science-based review of the European court’s decision on gene-edited crops. Nat. Biotechnol. 36, 800–802 (2018). & 2018.
Badran, A. H. & Liu, D. R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 6, 8425 (2015).
Cavaleiro, A. M., Kim, S. H., Seppälä, S., Nielsen, M. T. & Nørholm, M. H. H. Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth. Biol. 4, 1042–1046 (2015).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PloS ONE 3, e3647–e3647 (2008).
Lee, M. E., DeLoache, W. C., Cervantes, B. & Dueber, J. E. A. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 4, 975–986 (2015).
Potapov, V. et al. Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synth. Biol. 7, 2665–2674 (2018).
Ringquist, S. et al. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6, 1219–1229 (1992).
Davis, J. H., Rubin, A. J. & Sauer, R. T. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011).
Salis, H. M. The Ribosome Binding Site calculator. Method. Enzym. 498, 19–42 (2011).
Cui, L. et al. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9, 1912 (2018).
Chung, C. T. & Miller, R. H. Preparation and storage of competent Escherichia coli cells. Meth. Enzym. 218, 621–627 (1993).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotech. 37, 224–226 (2019).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Ashkenazy, H. et al. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Notredame, C., Higgins, D. G. & Heringa, J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Yang, J. & Zhang, Y. Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics 52, 5.8.1–5.8.15 (2015).
Acknowledgements
We thank B. Fu and C. Canavan for assistance with plasmid construction and assays; H. Rees, T. Wang, J. Bessen, A. Badran and P. Lichtor for helpful discussion; K. Clement for CRISPResso2 support; and A. Hamidi for help editing the manuscript. This work was supported by US NIH grant nos. U01 AI142756, RM1 HG009490, R01 EB022376 and R35 GM118062, St. Jude Collaborative Research Consortium, DARPA HR0011-17-2-0049, the Ono Pharma Foundation and HHMI. L.W.K. is an NSF Graduate Research Fellow and was supported by NIH Training Grant no. T32 GM095450. O.S.O. and J.R.H. were supported by NIH DC013521. C.Z. was supported by the Harvard College Research Program. C.W. is the Marion Abbe Fellow of the Damon Runyon Cancer Research Foundation (DRG-2343-18).
Author information
Authors and Affiliations
Contributions
B.W.T. designed the research, designed and constructed plasmids, and performed PACE and bacterial experiments. L.W.K. and J.M.L. designed and performed HEK cell experiments and analyzed data. J.M.L. designed and performed APOE editing experiments. W-H.Y. designed and performed baringo editing experiments. C.Z. constructed plasmids and performed bacterial experiments for selection development. G.A.N. designed and constructed plasmids for the HEK cell experiment for WFS1 editing. C.W. designed and performed ancestral sequence reconstruction. M.B., O.S-O. and J.R.H. contributed baringo mouse cells. D.R.L. designed and supervised the research. B.W.T., L.W.K. and D.R.L. wrote the manuscript. All authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.R.L. is a consultant and co-founder of Beam Therapeutics, Editas Medicine and Pairwise Plants, companies that use genome editing. D.R.L., B.W.T. and C.W. have filed patent applications on aspects of this work.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Complete Integrated SI
Supplementary Discussion 1–6, Supplementary Tables 1–2 and 4–6 and Supplementary Figs. 1–22
Supplementary Table 3
Compiled editing data/CRISPResso output tabulated for mammalian cell editing at C bases with measurable edits from any construct
Supplementary Data 1
Structure-guided alignment of APOBEC sequences used in Supplementary Fig. 17 in FASTA format
Supplementary Data 2
Homology model of APOBEC1
Supplementary Data 3
Homology model of FERNY
Supplementary Data 4
Homology model of CDA1
Rights and permissions
About this article
Cite this article
Thuronyi, B.W., Koblan, L.W., Levy, J.M. et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol 37, 1070–1079 (2019). https://doi.org/10.1038/s41587-019-0193-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0193-0
This article is cited by
-
Base-editing mutagenesis maps alleles to tune human T cell functions
Nature (2024)
-
CRISPR/Cas9-mediated base editors and their prospects for mitochondrial genome engineering
Gene Therapy (2024)
-
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Nature Communications (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Domain-inlaid Nme2Cas9 adenine base editors with improved activity and targeting scope
Nature Communications (2024)