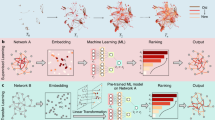
Abstract
Microbiomes from every environment contain a myriad of uncultivated archaeal and bacterial viruses, but studying these viruses is hampered by the lack of a universal, scalable taxonomic framework. We present vConTACT v.2.0, a network-based application utilizing whole genome gene-sharing profiles for virus taxonomy that integrates distance-based hierarchical clustering and confidence scores for all taxonomic predictions. We report near-identical (96%) replication of existing genus-level viral taxonomy assignments from the International Committee on Taxonomy of Viruses for National Center for Biotechnology Information virus RefSeq. Application of vConTACT v.2.0 to 1,364 previously unclassified viruses deposited in virus RefSeq as reference genomes produced automatic, high-confidence genus assignments for 820 of the 1,364. We applied vConTACT v.2.0 to analyze 15,280 Global Ocean Virome genome fragments and were able to provide taxonomic assignments for 31% of these data, which shows that our algorithm is scalable to very large metagenomic datasets. Our taxonomy tool can be automated and applied to metagenomes from any environment for virus classification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The set of reference genomes used to evaluate vConTACT was retrieved from https://www.ncbi.nlm.nih.gov/genome/viruses/. The GOV contigs were retrieved from the publicly available CyVerse data commons repository, accessible at http://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/GOV. The utility of vConTACT v.2.0 depends upon its expert evaluation and community availability39.
Code availability
The utility of vConTACT v.2.0 depends upon its expert evaluation and community availability. The tool is available through Bitbucket (https://bitbucket.org/MAVERICLab/vcontact2) as a downloadable Python package and usable as an app through iVirus39, the viral ecology apps and data resource embedded in the CyVerse Cyberinfrastructure, with detailed usage protocols available through Protocol Exchange (https://www.nature.com/protocolexchange/) and protocols.io (https://www.protocols.io/). Finally, the curated reference network is available at each of these sites and will be updated approximately bi-yearly as complete genomes become available and resources exist to support this effort.
References
Falkowski, P. G., Fenchel, T. & Delong, E. F. The microbial engines that drive earth’s biogeochemical cycles. Science 320, 1034–1039 (2008).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Moran, M. A. The global ocean microbiome. Science 350, aac8455 (2015).
Zhao, M. et al. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 8, 2045–2055 (2014).
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Fernández, L., Rodríguez, A. & García, P. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J. 12, 1171–1179 (2018).
Hurwitz, B. L. & U’Ren, J. M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol. 31, 161–168 (2016).
Suttle, C. A. Marine viruses – major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Brum, J. R. et al. Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015).
Danovaro, R. et al. Virus-mediated archaeal hecatomb in the deep seafloor. Sci. Adv. 2, e1600492 (2016).
Pratama, A. A. & van Elsas, J. D. The ‘neglected’ soil virome – potential role and impact. Trends Microbiol. https://doi.org/10.1016/j.tim.2017.12.004 (2018).
Gómez, P. & Buckling, A. Bacteria–phage antagonistic coevolution in soil. Science 332, 106–109 (2011).
Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F. & Gordon, J. I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10, 607–617 (2012).
Abeles, S. R. & Pride, D. T. Molecular bases and role of viruses in the human microbiome. J. Mol. Biol. 426, 3892–3906 (2014).
Rohwer, F. & Edwards, R. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184, 4529–4535 (2002).
Yarza, P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014).
Deng, L. et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242–245 (2014).
Gregory, A. C. et al. Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genomics 17, 930 (2016).
Bobay, L. & Ochman, H. Biological species in the viral world. PNAS 115, 6040–6045 (2018).
Mavrich, T. N. & Hatfull, G. F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2, 17112 (2017).
Ackermann, H.-W. in Methods and Protocols, Vol. 1 (eds Clokie, M. R. J. & Kropinski, A. M.) 127–140 (Humana Press, 2009).
Simmonds, P. et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161–168 (2017).
Paez-Espino, D. et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 47, D678–D686 (2018).
Brister, J. R., Ako-Adjei, D., Bao, Y. & Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 43, D571–D577 (2015).
Roux, S. et al. Minimum Information about an Uncultivated Virus Genome (MIUViG): a community consensus on standards and best practices for describing genome sequences from uncultivated viruses. Nat. Biotechnol. 37, 29–37 (2019).
Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379–2380 (2017).
Lima-Mendez, G., Van Helden, J., Toussaint, A. & Leplae, R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25, 762–777 (2008).
Bolduc, B. et al. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ 5, e3243 (2017).
Meier-Kolthoff, J. P. & Göker, M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33, 3396–3404 (2017).
Yu, C. et al. Real time classification of viruses in 12 dimensions. PLoS ONE 8, e64328 (2013).
Gao, Y. & Luo, L. Genome-based phylogeny of dsDNA viruses by a novel alignment-free method. Gene 492, 309–314 (2012).
Iranzo, J., Koonin, E. V., Prangishvili, D. & Krupovic, M. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J. Virol. 90, 11043–11055 (2016).
Aiewsakun, P. & Simmonds, P. The genomic underpinnings of eukaryotic virus taxonomy: creating a sequence-based framework for family-level virus classification. Microbiome 6, 38 (2018).
Lawrence, J. G., Hatfull, G. F. & Hendrix, R. W. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184, 4891–4905 (2002).
Lavigne, R. et al. Classification of myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9, 224 (2009).
Lavigne, R., Seto, D., Mahadevan, P., Ackermann, H. W. & Kropinski, A. M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159, 406–414 (2008).
Henz, S. R., Huson, D. H., Auch, A. F., Nieselt-Struwe, K. & Schuster, S. C. Whole-genome prokaryotic phylogeny. Bioinformatics 21, 2329–2335 (2005).
Iranzo, J., Krupovic, M. & Koonin, E. V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio 7, e00978-16 (2016).
Bolduc, B., Youens-Clark, K., Roux, S., Hurwitz, B. L. & Sullivan, M. B. IVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 11, 7–14 (2017).
Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016).
Vik, D. R. et al. Putative archaeal viruses from the mesopelagic ocean. PeerJ 5, e3428 (2017).
Roux, S. et al. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat. Commun. 8, 858 (2017).
Emerson, J. B. et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. https://doi.org/10.1038/s41564-018-0190-y (2018).
Martinez-Hernandez, F. et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 8, 15892 (2017).
De la Cruz Peña, M. J. et al. Deciphering the human virome with single-virus genomics and metagenomics. Viruses 10, 113 (2018).
Aiewsakun, P., Adriaenssens, E. M., Lavigne, R., Kropinski, A. M. & Simmonds, P. Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J. Gen. Virol. 99, 1331–1343 (2018).
Hulo, C., Masson, P., Le Mercier, P. & Toussaint, A. A structured annotation frame for the transposable phages: a new proposed family ‘Saltoviridae’ within the Caudovirales. Virology 477, 155–163 (2015).
Adriaenssens, E. M. et al. Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 163, 1125–1129 (2018).
Nepusz, T., Yu, H. & Paccanaro, A. Detecting overlapping protein complexes in protein–protein interaction networks. Nat. Methods 9, 471–472 (2012).
Doyle, E. L. et al. Genome sequences of four Cluster P Mycobacteriophages. Genome Announc. 6, e01101–e01117 (2018).
Pope, W. H. et al. Bacteriophages of Gordonia spp. display a spectrum of diversity and genetic relationships. MBio 8, e01069–17 (2017).
Pope, W. H. et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4, e06416 (2015).
Nelson, D. Phage taxonomy: we agree to disagree. J. Bacteriol. 186, 7029–7031 (2004).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996 (2018).
Krupovic, M., Quemin, E. R. J., Bamford, D. H., Forterre, P. & Prangishvili, D. Unification of the globally distributed spindle-shaped viruses of the archaea. J. Virol. 88, 2354–2358 (2014).
Rokyta, D. R., Burch, C. L., Caudle, S. B. & Wichman, H. A. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 188, 1134–1142 (2006).
Matsen, F. A., Kodner, R. B. & Armbrust, E. V. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 538 (2010).
Marz, M. et al. Challenges in RNA virus bioinformatics. Bioinformatics 30, 1793–1799 (2014).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Krupovic, M. et al. Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee. Arch. Virol. 161, 1095–1099 (2016).
Adams, M. J. et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 162, 2505–2538 (2017).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Wiwie, C., Baumbach, J. & Röttger, R. Comparing the performance of biomedical clustering methods. Nat. Methods 12, 1033–1038 (2015).
Brohée, S. & van Helden, J. Evaluation of clustering algorithms for protein–protein interaction networks. BMC Bioinformatics 7, 488 (2006).
Kamburov, A., Stelzl, U. & Herwig, R. IntScore: a web tool for confidence scoring of biological interactions. Nucleic Acids Res. 40, W140–W146 (2012).
Goldberg, D. S. & Roth, F. P. Assessing experimentally derived interactions in a small world. Proc. Natl Acad. Sci. USA 100, 4372–4376 (2003).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Ohio Supercomputer Center. http://osc.edu/ark:/19495/f5s1ph73 (1987).
Oliphant, T. E. SciPy: open source scientific tools for Python. Comput. Sci. Eng. 9, 10–20 (2007).
McKinney, W. Data structures for statistical computing in Python. in Proc. 9th Python Sci. Conf. 445, 51–56 (2010).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Csárdi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9 (2006).
Federico, P., Pfeffer, J., Aigner, W., Miksch, S. & Zenk, L. Visual analysis of dynamic networks using change centrality. in Proc. 2012 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 179–183 (IEEE, 2012).
Acknowledgements
We thank L. Bollinger, G. Trubl and I. Tolstoy for their comments on improving the manuscript, as well as Z.-Q. You for helping push the network analytics. High-performance computational support was provided as an award from the Ohio Supercomputer Center to M.B.S. Funding was provided in part by the Department of Energy’s Genome Sciences Program Soil Microbiome Scientific Focus Area award (no. SCW1632) to Lawrence Livermore National Laboratory, NSF Biological Oceanography awards (OCE no. 1536989 and OCE no. 1756314), and a Gordon and Betty Moore Foundation Investigator Award (no. 3790) to M.B.S. Funding was provided to J.R.B. by the Intramural Research Program of the National Institutes of Health (NIH) National Library of Medicine. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231 to S.R. This work was funded in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract no. HHSN272200700016I to J.H.K. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors.
Author information
Authors and Affiliations
Contributions
H.B.J., B.B. and M.B.S. designed the study. O.Z. and M.B.S. wrote the manuscript with substantial contributions from H.B.J., B.B., J.R.B., S.R., E.M.A., J.R.B., A.M.K., M.K., R.L. and D.T. H.B.J. and B.B. performed the statistical and network analyses.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Bipartite network of reference virus genomes generated by vConTACT v.2.0.
(a) The full network is represented with an oval indicating the area of close-up in (b) Close-up of 3 viral clusters displayed as bipartite network. In this configuration, pink nodes represent individual genomes, while dark gray nodes depict proteins clusters that are shared between viral clusters.
Supplementary Fig. 2 Clustering comparisons between vConTACT v.1.0 and v.2.0.
(a) Number of total viral clusters (VCs), and genus-assigned VCs, concordant and discordant VCs, as detected by vConTACT v.1.0 (left) and v.2.0 (right). Discordant VCs are (i) those that have a mix of the different genera (i.e., lumped genera), (ii) those that have different member virus(es) of the same genus split into multiple clusters (i.e., split genera), or (iii) mix of (i) and (ii). (b) Proteome similarities of viruses within 22 concordant VCs of vConTACT version 1.0. For all plots, the x-axis is the individual pairwise comparisons and y-axis is the proteome similarity (i.e., percentage fraction of shared protein clusters between genomes). VCs 6, 26, 66, and 130 (highlighted by bold borders) contain taxonomically-misplaced member virus(es) of the Che8virus, Pbunavirus, P100virus, and Bcep78virus, respectively, all of which have been correctly captured by v.2.0 and ratified by the ICTV (see Supplementary Fig. 5). Like these four VCs, the remaining 18 VCs contain distant relatives, with only 1–30% of similarities to the rest of the given clusters or discrete viral group(s). These 18 VCs display a number of discontinuous similarities, which were identified as outliers or separated VCs by v.2.0, respectively (see Supplementary Table 4).
Supplementary Fig. 3 vConTACT v.2.0-based detection and characterization of overlapping viral genomes.
(a) Box plots depicting the distribution of the topology-based confidence scores between viruses identified as overlaps (n = 74; min., 0.040; quartile (Q)1, 0.199; median, 0.340; Q3, 0.450; max., 0.623) and non-overlaps (n = 1,856 ; min., 0.000; Q1, 0.280; median, 0.499; Q3, 0.822; max., 1.000), which vConTACT v.2.0 placed into two clusters (see panel b) and single clusters, respectively. For details, see Methods. Comparison of topology-based confidence scores between overlaps and non-overlaps was performed by the one-sided Mann Whitney U test (P-value = 6.12e-09). (b) List of phages and archaeal viruses identified as overlaps and their ICTV genus. Details on the lifestyle and evolutionary modes of 74 viruses were collected from Mavrich and Hatfull22 and from the Actinobacteriophage Database website (http://phagesdb.org/). The high (HGCF, blue) and low (LGCF, green) gene content flux evolutionary modes indicate the predicted lifestyle based on the gene content dissimilarity between viral genomes. Bioinformatically-predicted temperate phages indicate those that contain integrase (for integrating temperate phage genomes into host) or parA (partitioning gene found in extrachromosomal temperate phages) genes.
Supplementary Fig. 4 Evaluation of optimal distance thresholds for hierarchical clustering of VCs.
The X-axis denotes distance threshold increments from dist = 1 to dist = 20 in 0.5 intervals. The Y-axis denotes composite scores by multiplying Accuracy (Acc) and clustering-wise separation (Sep) when trying to recapitulate ICTV genera, which are geometric means of Sensitivity and Positive predictive value, and Complex-wise separation and Cluster-wise separation, respectively. From these data, a distance of 9.0 yielded the highest composite score for the sub-clusters partitioned from all vConTACT v.2.0-generated viral clusters. For details, see Online Methods.
Supplementary Fig. 5 vConTACT v.2.0-based detection and characterization of boundary genome(s) within the ICTV-recognized genera.
(a) Box plots show the percentage of shared protein clusters (PCs) between members of an ICTV genus (red), and the same metrics after excluding viruses recognized as outlier(s) by vConTACT v.2.0 (cyan). The proteome similarities for the Barnyardvirus are shown between (1) member viruses of the Barnyardvirus, (2) Mycobacterium virus Barnyard and the remaining members of the Barnyardvirus, (3) the Barnyardvirus and Patiencevirus, (4) Mycobacterium virus Barnyard and the Patiencevirus, and (5) the Barnyardvirus without outliers. For the Phikmvvirus, the proteome similarities between (1) member viruses of the Phikmvvirus, (2) Pseudomonas virus phiKMV and the remaining members of the Phikmvvirus, and (3) Pseudomonas virus phiKMV and members of VC33 are shown, respectively. The sample size (n) as well as minimum (min.), median, maximum (max.) and two quartiles (Q1 and Q3) for each plot were represented as follows. From left box plot to right, (n = 5; min., 27.1; Q1, 27.7; median, 88.1; Q3, 90.4; max., 93.5), (4; 88.0; 88.5; 90.0; 91.2; 93.5), (10; 24.0; 29.3; 83.3; 91.0; 97.7), (8; 76.5; 84.8; 90.1; 92.8; 97.7), (11; 36.1; 91.9; 93.9; 95.5; 97.8), (10; 90.5; 93.3; 94.4; 95.6; 97.8), (5; 49.4; 51.3; 85.9; 86.7; 97.6), (4; 85.8; 86.1; 86.6; 92.9; 97.6), (17; 25.7; 59.3; 67.3; 72.9; 94.3), (15;58.0; 65.3; 69.0;74.1; 94.3), (2; 65.7; 65.7; 65.7; 65.7; 65.7), (10; 58.4; 76.5; 81.5; 84.1; 94.1); (9; 74.3; 80.3; 83.0; 84.6; 94.1), (31; 12.8; 35.9; 44.1; 57.6; 92.4), (25; 29.9; 42.3; 50.0; 63.1; 92.4), (5; 16.0; 16.8; 20.3; 56.5; 66.7), (3; 64.8; 65.7; 66.7; 66.7; 66.7), (2; 31.6; 31.6; 31.6; 31.6; 31.6), (3; 43.5; 43.7; 43.9; 60.7; 77.4), (3; 43.5; 43.6; 43.7; 43.8; 43.9), (4; 42.6; 43.5; 43.9; 44.1; 77.4), (3; 38.7; 38.7; 38.7; 38.7; 38.7; 38.7), (3; 77.4; 77.4; 77.4; 77.4; 77.4; 77.4), (4; 19.3; 21.6; 27.9; 29.0; 45.8), (3; 19.3; 23.1; 27.0; 36.4; 45.8), (11; 85.5; 89.0; 90.3; 93.1; 95.2). (b) Module profiles show the presence (dark) and absence (light) of homologous PCs across genomes. Each row represents a virus and each column a PC. The genomes were hierarchically clustered based on pairwise Euclidean distance. The ICTV and vConTACT v.2.0 classifications are indicated next to each virus.
Supplementary Fig. 6 Evaluation of singletons/outlier genomes over GOV increments.
Thirty-eight ICTV recognized singleton and outlier genomes (one per row) were observed to evaluate whether the addition of GOV sequences would improve their classification. The coloring gradient on the left indicates the numbers of genera per VC. Clearly improved clustering was observed in 3 of the 38 genomes (black bolded, left), with 2 genomes clustering to 6-genera discordant VCs (red, left), though the majority saw only minor, if any, change in their clustering assignment.
Supplementary Fig. 7 vConTACT v.2.0 computational runtimes.
In the plot, processing time (in seconds, Y-axis) is plotted against increasing GOV sequence data (GOV % added, X-axis), where the number of GOV contigs per additional increment is placed in parentheses on the X-axis. There is a strong linear correlation between runtime and memory usage with data volume to be processed. The R2 value, or coefficient of determination, was calculated as [1 - residual sum of squares / total sum of squares].
Supplementary Fig. 8 Impact of the inflation factor on viral genome clustering based on the Markov clustering (MCL) algorithm.
Left panel: average intra-cluster clustering coefficient (ICCC) and number of viral clusters (VCs), which are predicted as a function of the inflation factors ranging from 1.0 to 5.0 with a step of 0.2, are indicated. Right panel: Curve representing the ICCC values for the network containing 2,304 archaeal and bacterial virus genomes.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8 and Supplementary Notes 1 and 2
Supplementary Table 1
A genome–gene matrix.
Supplementary Table 2
List of 2,304 archaeal and bacterial virus genomes used to evaluate vConTACT v.1.0 and v.2.0.
Supplementary Table 3
Clustering performance evaluations of the vConTACT v.1.0, v.2.0, and v.2.0 followed by distance-based hierarchical clustering for the genus rank.
Supplementary Table 4
Fraction of PCs in common between 2,304 genomes.
Supplementary Table 5
Statistics associated with the box plots shown in Fig. 2d.
Supplementary Table 6
Statistics associated with the box plots shown in Fig. 3b.
Rights and permissions
About this article
Cite this article
Bin Jang, H., Bolduc, B., Zablocki, O. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37, 632–639 (2019). https://doi.org/10.1038/s41587-019-0100-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0100-8
This article is cited by
-
Comparative analysis of the vaginal bacteriome and virome in healthy women living in high-altitude and sea-level areas
European Journal of Medical Research (2024)
-
Diversity and potential host-interactions of viruses inhabiting deep-sea seamount sediments
Nature Communications (2024)
-
Bacteriophages from human skin infecting coagulase-negative Staphylococcus: diversity, novelty and host resistance
Scientific Reports (2024)
-
Characterization of the gut bacterial and viral microbiota in latent autoimmune diabetes in adults
Scientific Reports (2024)
-
A metagenomic catalog of the early-life human gut virome
Nature Communications (2024)