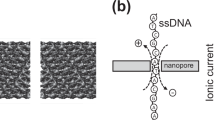
Abstract
Nanopore DNA sequencing is limited by low base-calling accuracy. Improved base-calling accuracy has so far relied on specialized base-calling algorithms, different nanopores and motor enzymes, or biochemical methods to re-read DNA molecules. Two primary error modes hamper sequencing accuracy: enzyme mis-steps and sequences with indistinguishable signals. We vary the driving voltage from 100 to 200 mV, with a frequency of 200 Hz, across a Mycobacterium smegmatis porin A (MspA) nanopore, thus changing how the DNA strand moves through the nanopore. A DNA helicase moves the DNA through the nanopore in discrete steps, and the variable voltage moves the DNA continuously between these steps. The electronic signal produced with variable voltage is used to overcome the primary error modes in base calling. We found that single-passage de novo base-calling accuracy of 62.7 ± 0.5% with a constant driving voltage improves to 79.3 ± 0.3% with a variable driving voltage. The variable-voltage sequencing mode is complementary to other methods to boost the accuracy of nanopore sequencing and could be incorporated into any enzyme-actuated nanopore sequencing device.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data for the main text figures as well as for all the constant- and variable-voltage sequencing reads used for the validation study can be found on figshare at https://doi.org/10.6084/m9.figshare.7723214. This also contains all of the Matlab code and supporting files that are necessary to replicate thesequencing analysis for both constant- and variable-voltage, as well as the scripts to generate the main text figures from their underlying data.
Code availability
Code and supporting files for constant- and variable-voltage sequencing analysis, as well as for main text figure generation can be found on github at https://github.com/uwnanopore/variable-voltage-sequencing.git.
References
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. 93, 13770–13773 (1996).
Manrao, E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012).
Laszlo, A. H. et al. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 32, 829–833 (2014).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Craig, J. M. et al. Direct detection of unnatural DNA nucleotides dNaM and d5SICS using the MspA nanopore. PLoS ONE 10, e0143253 (2015).
Laszlo, A. H. et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl Acad. Sci. 110, 18904–18909 (2013).
Schreiber, J. et al. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc. Natl Acad. Sci. USA 110, 18910–18915 (2013).
Rand, A. C. et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153 (2008).
Castro-Wallace, S. L. et al. Nanopore DNA sequencing and genome assembly on the International Space Station. Sci. Rep. 7, 18022 (2017).
Loman, N. J., Quick, J. & Simpson, J. T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735 (2015).
Theuns, S. et al. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Sci. Rep. 8, 9830 (2018).
Bronzato Badial, A. et al. Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis. 1648–1652 (2018).
Cao, M. D. et al. Streaming algorithms for identification pathogens and antibiotic resistance potential from real-time MinIONTM sequencing. GigaScience 5, 1–16 (2016).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Quick, J. et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016).
Quick, J. et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 16, 114 (2015).
Faria, N. R. et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546, 406–410 (2017).
Kafetzopoulou, L. E. et al. Assessment of Metagenomic MinION and Illumina sequencing as an approach for the recovery of whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Preprint at https://doi.org/10.1101/355560 (2018).
Pomerantz, A. et al. Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. GigaScience 7, giy033 (2018).
Cherf, G. M. et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat. Biotechnol. 30, 344–348 (2012).
Derrington, I. M. et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 33, 1073–1075 (2015).
Jain, M. et al. MinION analysis and reference consortium: phase 2 data release and analysis of R9.0 chemistry. F1000Res. 6, 760 (2017).
Rang, F. J., Kloosterman, W. P. & de Ridder, J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19, 90 (2018).
Bhattacharya, S. et al. Molecular dynamics study of MspA arginine mutants predicts slow DNA translocations and ion current blockades indicative of DNA sequence. ACS Nano 6, 6960–6968 (2012).
Craig, J. M. et al. Revealing dynamics of helicase translocation on single-stranded DNA using high-resolution nanopore tweezers. Proc. Natl Acad. Sci. 114, 11932–11937 (2017).
Butler, T. Z., Pavlenok, M., Derrington, I. M., Niederweis, M. & Gundlach, J. H. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl Acad. Sci. 105, 20647–20652 (2008).
Manrao, E. A., Derrington, I. M., Pavlenok, M., Niederweis, M. & Gundlach, J. H. Nucleotide discrimination with DNA immobilized in the MspAnanopore. PLoS ONE 6, e25723 (2011).
Krishnakumar, R. et al. Systematic and stochastic influences on the performance of the MinION nanopore sequencer across a range of nucleotide bias. Sci. Rep. 8, 3159 (2018).
Rames, E. & Macdonald, J. Evaluation of MinION nanopore sequencing for rapid enterovirus genotyping. Virus Res. 252, 8–12 (2018).
Boža, V., Brejová, B. & Vinař, T. DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS ONE 12, e0178751 (2017).
Teng, H. et al. Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience 7, giy037 (2018).
David, M. et al. Nanocall: an open source basecaller for Oxford Nanopore sequencing data. Bioinformatics 33, 49–55 (2017).
Vaser, R., Sovic, I., Nagarajan, N. & Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. https://doi.org/10.1101/gr.214270.116 (2017).
Simpson, J. Signal-level algorithms for MinION data. Github https://github.com/jts/nanopolish (2018).
Laszlo, A. H., Derrington, I. M. & Gundlach, J. H. MspA nanopore as a single-molecule tool: from sequencing to SPRNT. Methods 105, 75–89 (2016).
Acknowledgements
This work was supported by the National Human Genome Research Institute grant R01HG005115 (to J.H.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors J.H.G., M.T.N., and H.B., along with the University of Washington, have filed provisional patent applications covering the methods presented in this work. The patent has been filed under application number 62/805,870 by the University of Washington CoMotion.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–19, Supplementary Tables 1–3 and Supplementary Notes 1–15
Rights and permissions
About this article
Cite this article
Noakes, M.T., Brinkerhoff, H., Laszlo, A.H. et al. Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nat Biotechnol 37, 651–656 (2019). https://doi.org/10.1038/s41587-019-0096-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0096-0
This article is cited by
-
Detection of ribonucleotides embedded in DNA by Nanopore sequencing
Communications Biology (2024)
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
Metagenomic surveillance for bacterial tick-borne pathogens using nanopore adaptive sampling
Scientific Reports (2023)
-
Detection of phosphorylation post-translational modifications along single peptides with nanopores
Nature Biotechnology (2023)
-
Nanopore Sequencing Discloses Compositional Quality of Commercial Probiotic Feed Supplements
Scientific Reports (2023)