Abstract
In many species, including mice, female animals show markedly different pup-directed behaviours based on their reproductive state1,2. Naive wild female mice often kill pups, while lactating female mice are dedicated to pup caring3,4. The neural mechanisms that mediate infanticide and its switch to maternal behaviours during motherhood remain unclear. Here, on the basis of the hypothesis that maternal and infanticidal behaviours are supported by distinct and competing neural circuits5,6, we use the medial preoptic area (MPOA), a key site for maternal behaviours7,8,9,10,11, as a starting point and identify three MPOA-connected brain regions that drive differential negative pup-directed behaviours. Functional manipulation and in vivo recording reveal that oestrogen receptor α (ESR1)-expressing cells in the principal nucleus of the bed nucleus of stria terminalis (BNSTprESR1) are necessary, sufficient and naturally activated during infanticide in female mice. MPOAESR1 and BNSTprESR1 neurons form reciprocal inhibition to control the balance between positive and negative infant-directed behaviours. During motherhood, MPOAESR1 and BNSTprESR1 cells change their excitability in opposite directions, supporting a marked switch of female behaviours towards the young.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Fibre photometry recording data, behaviour annotations and raw representative histology images are available at Zenodo (https://doi.org/10.5281/zenodo.7772552). Behaviour videos and additional histology images are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
MATLAB code used in this study is available at Zenodo (https://doi.org/10.5281/zenodo.7772552).
References
Parmigiani, S. & vom Saal, F. Infanticide And Parental Care (Routledge, 2016).
Dulac, C., O’Connell, L. A. & Wu, Z. Neural control of maternal and paternal behaviors. Science 345, 765–770 (2014).
Soroker, V. & Terkel, J. Changes in incidence of infanticidal and parental responses during the reproductive cycle in male and female wild mice Mus musculus. Anim. Behav. 36, 1275–1281 (1988).
McCarthy, M. M. & vom Saal, F. S. The influence of reproductive state on infanticide by wild female house mice (Mus musculus). Physiol. Behav. 35, 843–849 (1985).
Kohl, J., Autry, A. E. & Dulac, C. The neurobiology of parenting: a neural circuit perspective. Bioessays 39, 1–11 (2017).
Numan, M. Motivational systems and the neural circuitry of maternal behavior in the rat. Dev. Psychobiol. 49, 12–21 (2007).
Kohl, J. & Dulac, C. Neural control of parental behaviors. Curr. Opin. Neurobiol. 49, 116–122 (2018).
Yu, Z.-X., Li, X.-Y. & Xu, X.-H. Neural circuit mechanisms that underlie parental care. Adv. Exp. Med. Biol. 1284, 49–62 (2020).
Numan, M. & Insel, T. R. The Neurobiology of Parental Behavior (Springer, 2003).
Kuroda, K. O. & Numan, M. The medial preoptic area and the regulation of parental behavior. Neurosci. Bull. 30, 863–865 (2014).
Carollo, A., Balagtas, J. P. M., Neoh, M. J. & Esposito, G. A scientometric approach to review the role of the medial preoptic area (MPOA) in parental behavior. Brain Sci. 11, 393 (2021).
Lukas, D. & Huchard, E. The evolution of infanticide by females in mammals. Philos. Trans. R. Soc. B 374, 20180075 (2019).
Palombit, R. A. Infanticide as sexual conflict: coevolution of male strategies and female counterstrategies. Cold Spring Harb. Perspect. Biol. 7, a017640 (2015).
Jakubowski, M. & Terkel, J. Infanticide and caretaking in non-lactating Mus musculus: influence of genotype, family group and sex. Anim. Behav. 30, 1029–1035 (1982).
Svare, B. & Mann, M. Infanticide: genetic, developmental and hormonal influences in mice. Physiol. Behav. 27, 921–927 (1981).
Gandelman, R. The ontogeny of maternal responsiveness in female Rockland-Swiss albino mice. Horm. Behav. 4, 257–268 (1973).
Wei, Y. C. et al. Medial preoptic area in mice is capable of mediating sexually dimorphic behaviors regardless of gender. Nat. Commun. 9, 279 (2018).
Fang, Y. Y., Yamaguchi, T., Song, S. C., Tritsch, N. X. & Lin, D. A hypothalamic midbrain pathway essential for driving maternal behaviors. Neuron 98, 192–207 (2018).
Wu, Z., Autry, A. E., Bergan, J. F., Watabe-Uchida, M. & Dulac, C. G. Galanin neurons in the medial preoptic area govern parental behaviour. Nature 509, 325–330 (2014).
Kohl, J. et al. Functional circuit architecture underlying parental behaviour. Nature 556, 326–331 (2018).
Tsuneoka, Y. et al. Neurotransmitters and neuropeptides in gonadal steroid receptor-expressing cells in medial preoptic area subregions of the male mouse. Sci. Rep. 7, 9809 (2017).
Li, X. Y. et al. AGRP neurons project to the medial preoptic area and modulate maternal nest-building. J. Neurosci. 39, 456–471 (2019).
Zhang, G.-W. et al. Medial preoptic area antagonistically mediates stress-induced anxiety and parental behavior. Nat. Neurosci. 24, 516–528 (2021).
Tsuneoka, Y. et al. Distinct preoptic-BST nuclei dissociate paternal and infanticidal behavior in mice. EMBO J. 34, 2652–2670 (2015).
Autry, A. E. et al. Urocortin-3 neurons in the mouse perifornical area promote infant-directed neglect and aggression. eLife 10, e64680 (2021).
Sato, K. et al. Amygdalohippocampal area neurons that project to the preoptic area mediate infant-directed attack in male mice. J. Neurosci. 40, 3981–3994 (2020).
Chen, P. B. et al. Sexually dimorphic control of parenting behavior by the medial amygdala. Cell 176, 1206–1221 (2019).
Numan, M. Medial preoptic area and maternal behavior in the female rat. J. Compar. Physiol. Psychol. 87, 746–759 (1974).
Numan, M., Corodimas, K. P., Numan, M. J., Factor, E. M. & Piers, W. D. Axon-sparing lesions of the preoptic region and substantia innominata disrupt maternal behavior in rats. Behav. Neurosci. 102, 381–396 (1988).
Mann, M. A., Kinsley, C., Broida, J. & Svare, B. Infanticide exhibited by female mice: genetic, developmental and hormonal influences. Physiol. Behav. 30, 697–702 (1983).
Zingg, B. et al. AAV-mediated anterograde transsynaptic tagging: mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron 93, 33–47 (2017).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Chen, A.-X. et al. Specific hypothalamic neurons required for sensing conspecific male cues relevant to inter-male aggression. Neuron 108, 763–774 (2020).
Stagkourakis, S. et al. A neural network for intermale aggression to establish social hierarchy. Nat. Neurosci. 21, 834–842 (2018).
Gallo, M. et al. Limited bedding and nesting induces maternal behavior resembling both hypervigilance and abuse. Front. Behav. Neurosci. 13, 167 (2019).
Nephew, B. C. & Bridges, R. S. Effects of chronic social stress during lactation on maternal behavior and growth in rats. Stress 14, 677–684 (2011).
Kim, S. Y. et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature 496, 219–223 (2013).
Jennings, J. H. et al. Distinct extended amygdala circuits for divergent motivational states. Nature 496, 224–228 (2013).
Lebow, M. A. & Chen, A. Overshadowed by the amygdala: the bed nucleus of the stria terminalis emerges as key to psychiatric disorders. Mol. Psychiatry 21, 450–463 (2016).
Mitra, S. W. et al. Immunolocalization of estrogen receptor β in the mouse brain: comparison with estrogen receptor α. Endocrinology 144, 2055–2067 (2003).
Lee, H. et al. Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature 509, 627–632 (2014).
Hashikawa, K. et al. Esr1+ cells in the ventromedial hypothalamus control female aggression. Nat. Neurosci. 20, 1580–1590 (2017).
Yamaguchi, T. et al. Posterior amygdala regulates sexual and aggressive behaviors in male mice. Nat. Neurosci. 23, 1111–1124 (2020).
Kelly, D. A., Varnum, M. M., Krentzel, A. A., Krug, S. & Forger, N. G. Differential control of sex differences in estrogen receptor α in the bed nucleus of the stria terminalis and anteroventral periventricular nucleus. Endocrinology 154, 3836–3846 (2013).
Nguyen, A. Q., Dela Cruz, J. A., Sun, Y., Holmes, T. C. & Xu, X. Genetic cell targeting uncovers specific neuronal types and distinct subregions in the bed nucleus of the stria terminalis. J. Comp. Neurol. 524, 2379–2399 (2016).
Hrdy, S. B. Infanticide among animals: a review, classification, and examination of the implications for the reproductive strategies of females. Ethol. Sociobiol. 1, 13–40 (1979).
Knoedler, J. R. et al. A functional cellular framework for sex and estrous cycle-dependent gene expression and behavior. Cell 185, 654–671 (2022).
Yang, B., Karigo, T. & Anderson, D. J. Transformations of neural representations in a social behaviour network. Nature 608, 741–749 (2022).
Zhou, X. et al. Hyperexcited limbic neurons represent sexual satiety and reduce mating motivation. Science 379, 820–825 (2023).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Yin, L. et al. VMHvllCckar cells dynamically control female sexual behaviors over the reproductive cycle. Neuron 110, 3000–3017 (2022).
Wong, L. C. et al. Effective modulation of male aggression through lateral septum to medial hypothalamus projection. Curr. Biol. 26, 593–604 (2016).
Falkner, A. L. et al. Hierarchical representations of aggression in a hypothalamic-midbrain circuit. Neuron 106, 637–648 (2020).
Acknowledgements
We thank all of the members of the Lin laboratory for discussions; and Y. Jiang for assisting with genotyping. This research was supported by NIH grants R01MH101377, R01MH124927 and U19NS107616 (to D.L.), R01HD092596 (to D.L. and R.M.S.), and R37HD083217 (to R.M.S.); the Mathers Foundation, and the Vulnerable Brain Project (to D.L.); and the Levy Leon Postdoctoral Fellowship (to L.M.).
Author information
Authors and Affiliations
Contributions
D.L. and L.M. conceived the project, designed experiments, analysed data and wrote the paper. D.L. supervised the project. L.M. performed most of the experiments and prepared figures. R.Y. and L.Y. performed slice recording experiments and prepared related figures. R.M.S. provided critical feedback on the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Minmin Luo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
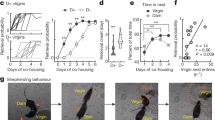
Extended Data Fig. 1 Ablation or chemogenetic inhibition of MPOAEsr1 cells induces infanticide in SW virgin female mice.
(a) Experimental design to ablate MPOAEsr1 cells. (b) Experimental timeline. (c,d) Left showing representative histology images of Esr1 staining (magenta) in MPOA and BNSTpr after DT injection in a mCherry female of 6 mice(c) and a DTR female of 8 mice (d). Right showing raster plots of pup-directed behaviours in mCherry (c) and DTR females (d) before and after i.p. injection of DT. *Remove wounded pups and stop recording. (e) Number of mCherry and DTR virgin females that attack, ignore or retrieve pups before and after DT injection. Fisher’s exact test for comparing behaviours (attack vs. no attack) between groups before or after DT injection. McNemar’s test for comparing behaviours (attack vs. no attack) between pre-DT and after-DT within a group. * p < 0.05. **p < 0.01. (f,g) Latency to attack pup (f) and investigate pup (g) before and after DT injection in mCherry and DTR females. If the behaviour of interest is not observed during the entire session, the latency is 600 s. Error bars: ± SEM. Mixed-effects analysis followed with multiple comparisons test. ****p < 0.0001. n = 6 mice for mCherry group, n = 8 mice for DTR group. (h) Experimental design to chemogenetically inhibit MPOAEsr1 cells. (i) Experimental timeline. (j,k) Left showing representative histology images of mCherry (j) and hM4Di-mCherry (k) expression of 6 mice each group in MPOA. Right showing raster plots of pup-directed behaviours in mCherry (j) and hM4Di females (k) after i.p. injection of saline or CNO. *Remove wounded pups and stop recording. (l) Number of mCherry and hM4Di virgin females that attack, ignore or retrieve pups after saline or CNO injection. Fisher’s exact test for comparing behaviours (attack vs. no attack) between groups after saline or CNO injection. McNemar’s test for comparing behaviours (attack vs. no attack) between saline and CNO injections within a group. * p < 0.05. **p < 0.01. (m,n) Latency to attack pup (m) and investigate pup (n) after saline or CNO injection in mCherry and hM4Di females. Error bars: ± SEM. Mixed-effects analysis (m) and Two-way RM ANOVA (n) followed with multiple comparisons test. ****p < 0.0001. n = 6 mice for each group. Brain illustrations in (a) and (h) are produced based on reference atlas from https://atlas.brain-map.org/.See Source Data Extended Data Fig. 1 for detailed values and statistics.
Extended Data Fig. 2 MPOA-connecting cells across brain regions and their overlap with Infanticide and maternal care induced c-Fos.
(a) Experimental design to trace MPOA-connecting cells throughout the brain using Ai6 female mice and high titre (> 1 × 10¹³ vg/mL) AAV1-hSyn-Cre. (b and c) Images from a representative animal of 4 mice showing the primary injection site in the MPOA (b) and MPOA-connecting cells in various brain areas (c). Scale bars: 1 mm (b) and 200 µm (c). (d) Distribution of MPOA-connecting neurons in various brain regions. All regions containing over 1% of total labelled cells are shown. n = 4. Error bars: SEM. (e–g) Representative images of 3 mice each group of 18 regions showing baseline (e), infanticide-induced (f), and maternal behaviour-induced (g) c-Fos and zsGreen expression in Ai6 female mice injected with AAV1-hSyn-Cre into the MPOA. Baseline c-Fos are from females left undisturbed in the home cage. Brain illustrationsare produced based on reference atlas from https://atlas.brain-map.org/. (h) The density of c-Fos expressing cells in each MPOA-connecting brain region in control, infanticidal and maternal female mice. n = 3 mice for each group. One-way ANOVA followed with multiple comparisons test. *p < 0.05, **p < 0.01, Error bars: ±SEM. (i) The total number of Fos+Tracer+ and Fos-Trace+ cells per 100 Tracer+ cells in each MPOA-connecting brain region in control, infanticidal and maternal female mice. n = 3 mice for each group. Fisher’s exact test is based on unnormalized total numbers of Fos+Tracer+ and Fos-Trace+ cells of each group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See Source Data Extended Data Fig. 2 for detailed values and statistics.
Extended Data Fig. 3 Pup-directed behaviours after chemogenetic activation of MPOA-connecting cells in the VMHvl, SUM, PVT, PVN, or PMv in female mice.
(a) Experimental design to chemogenetically activate various MPOA-connecting regions. (b) Experimental timeline. (c1, d1, e1, f1, g1) Representative histology images of 8 mice showing hM3Dq-mCherry expression in the VMHvl (c1), SUM (d1), PVT (e1), PVN (f1), and PMv (g1) after injecting AAV1-Syn-Cre in MPOA and AAV2-hSyn-DIO-hM3Dq-mCherry into each of the regions. Scale bars: 200 µm. (c2, d2, e2, f2, g2) Representative raster plots showing pup-directed behaviours after saline or CNO injection into animals expressing hM3Dq-mCherry in MPOA-connecting cells in the VMHvl (c2), SUM (d2), PVT (e2), PVN (f2), and PMv (g2). Each raster lasts 10 min. (c3, d3, e3, f3, g3) Pup-directed attack after saline or CNO injection in female mice that express hM3Dq-mCherry in MPOA-connecting cells in the VMHvl (c3), SUM (d3), PVT (e3), PVN (f3), and PMv (g3). Each circle represents one mouse. n = 8 for each group. (c4, d4, e4, f4, g4) Duration of pup investigation between saline- and CNO-injected days in female mice that express hM3Dq-mCherry in MPOA-connecting cells in the VMHvl (c4), SUM (d4), PVT (e4), PVN (f4), and PMv (g4). Each grey line represents one animal. The coloured line represents the group average. Error bars: ± SEM. Wilcoxon matched-pairs signed rank test (c4) or Two-tailed Paired t test (d4, e4, f4, g4). **p < 0.01. n = 8 for each group. (c5, d5, e5, f5, g5) Duration of pup grooming between saline- and CNO-injected days in female mice that express hM3Dq-mCherry in MPOA-connecting cells in the VMHvl (c5), SUM (d5), PVT (e5), PVN (f5), and PMv (g5). Figure conventions as in c4-g4. (c6, d6, e6, f6, g6) Latency to first pup attack between saline- and CNO-injected days in female mice that express hM3Dq-mCherry in MPOA-connecting cells in the VMHvl (c6), SUM (d6), PVT (e6), PVN (f6), and PMv (g6). Figure conventions as in c4-g4. See Source Data Extended Data Fig. 3 for detailed values and statistics.
Extended Data Fig. 4 Optogenetic activation of BNSTprMPOA neurons elicits infanticide, induces real-time place preference, and reduces anxiety in female mice.
(a) Experimental design to optogenetically activate BNSTprMPOA cells. (b) Experimental timeline. (c) Light delivery protocol. (d and f) Representative raster plots showing pup-directed behaviours during sham and 2 mW light stimulation in virgin female mice expressing GFP (d) or ChR2-EYFP (f) in BNSTprMPOA cells. # Remove wounded pup(s) and introduce a new pup. (e and g) PETH of attack pup probability in virgin female mice expressing GFP (e) and virgin female mice expressing ChR2-EYFP (g) in BNSTprMPOA cells following sham or light stimulation. Only trials with female-pup contact are included for analysis. Left black dash line: Light on. Right red dash line: light off. (h) Percentage of animals that attacked pups in GFP and ChR2 group. Fisher’s exact test for comparison between GFP and ChR2 group. McNemar’s test for comparison between sham and different laser intensity within ChR2 group. *p < 0.05, **p < 0.01, ****p < 0.0001. n = 8 mice for GFP control, n = 11 mice for ChR2 virgin group (sham, 0-1, 2-3), n = 10 mice for ChR2 virgin group (>3). (i) Percentage of trials showing pup attack. Each dot represents one mouse. Error bars: ± SEM. Mixed-effects analysis followed with multiple comparisons test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n = 8 mice for GFP control, n = 11 mice for ChR2 virgin group (sham, 0-1, 2-3), n = 10 mice for ChR2 virgin group (>3). (j) Averaged latency to attack pup upon encountering the pup following sham or light stimulation. Error bars: ± SEM. Mixed-effects analysis followed with multiple comparisons test. ****p < 0.0001. n = 8 mice for GFP control, n = 11 mice for ChR2 virgin group (sham, 0-1, 2-3), n = 10 mice for ChR2 virgin group (>3). (k-l) Representative tracking results during the RTPP test (k) and the time spent in sham or 3 mW stimulation chamber (l). n = 8 mice. Paired t test. *p < 0.05. Error bars: SEM. (m-n) Representative tracking results during an EPM test (m) and the time spent in open arms (n) with sham or 3 mW laser stimulation. n = 8 mice. Paired t test. *p < 0.05. Error bars: SEM. See Source Data Extended Data Fig. 4 for detailed values and statistics.
Extended Data Fig. 5 Additional characterization of BNSTpr Esr1 cells and their activation-induced infanticide in SW female mice.
(a) Representative coronal sections of 3 mice each group showing Esr1 immunostaining in MPOA and BNSTpr in a male (top), a virgin female (middle) and a mother (bottom). Scale bars: 500 µm. (b, c) Number of Esr1 positive cells in the MPOA (b) and BNSTpr (c) in male, virgin female, and mother mice. n = 3 mice for each group. One-way ANOVA followed with multiple comparisons test. **p < 0.01, ****p < 0.0001. Error bars: ± SEM. (d) Representative images of 3 mice showing overlap between Esr1 (red) and infanticide-induced c-Fos (green) in the BNSTpr. Right shows the enlarged view of the boxed area. Scale bars: 100 µm (left) and 10 µm (right). (e) Representative images of 3 mice showing the overlap between Esr1 (red) and zsGreen (green) in the BNSTpr in an Ai6 animals injected with AAV1-hSyn-Cre at the MPOA. Right shows the enlarged view of the boxed area. Scale bars: 100 µm (left) and 10 µm (right). (f) Quantification of overlap between MPOA-connected cells, Esr1 and the infanticide-induced c-Fos in the BNSTpr. n = 3 mice for each group, Error bars: ± SEM. (g) Experimental design to optogenetically activate BNSTprEsr1 cells in non-infanticidal SW virgin females. (h) Experimental timeline. (i) Representative image of 4 mice showing ChR2 expression (green) in the BNSTpr and fibre tracks (while boxes). Scale bar: 1 mm. (j) Representative raster plots showing pup-directed behaviours during sham and 2 mW light stimulation in a non-infanticidal SW virgin female (top) and a mother (bottom) expressing ChR2-EYFP in BNSTprEsr1 cells. # Replace a wounded pup. (k) Percentage of animals that attack pups. n = 5 mice for ChR2 virgin group, n = 4 mice for ChR2 mother group. McNemar’s test for comparison between sham and different laser intensity within each ChR2 group. (l) Percentage of trials showing pup attack. Each dot represents one mouse. Mean ± SEM. Mixed-effects analysis followed with multiple comparisons test. **p < 0.01, ***p < 0.001, ****p < 0.0001. n = 5 mice for ChR2 virgin group, n = 4 mice for ChR2 mother group. (m) Average latency to attack pup upon encountering the pup following sham or light stimulation. Error bars: ± SEM. Mixed-effects analysis followed with multiple comparisons test. ***p < 0.001, ****p < 0.0001. n = 5 mice for ChR2 virgin group, n = 4 mice for ChR2 mother group. See Source Data Extended Data Fig. 5 for detailed values and statistics.
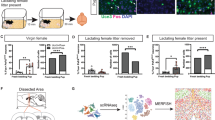
Extended Data Fig. 6 Chemogenetic inhibition of BNSTprEsr1 neurons promotes maternal behaviour in non-hostile virgin females and mothers.
(a) Representative raster plots showing pup-directed behaviours in non-hostile non-maternal mCherry and hM4Di virgin females after saline or CNO injection. (b) Percentage of mCherry and hM4Di virgin females that ignore, retrieve or attack pups after saline or CNO injection. Fisher’s exact test between mCherry and hM4Di group. (c,d) Latency to retrieve pup (c) and pup investigation duration (d) after saline or CNO injection in non-hostile non-maternal virgin mCherry and hM4Di females. Error bars: ± SEM. n = 6 mice for mCherry group; n = 5 mice for hM4Di group. Mixed-effects analysis followed with multiple comparisons test. **p < 0.01. (e) Representative raster plots showing various pup-directed behaviours in lactating mCherry and hM4Di females after saline or CNO injection. (f) All mCherry and hM4Di lactating females retrieved pups after saline or CNO injection. (g) All 5 pups were retrieved in the 10-min testing period in mCherry and hM4Di lactating females after either saline or CNO injection. n = 9 mice for mCherry group; n = 11 mice for hM4Di group. (h) Latency to retrieve the first pup and all five pups in mCherry and hM4Di lactating females after saline or CNO injection. Error bars: SEM. n = 9 mice for mCherry group; n = 11 mice for hM4Di group. Mixed-effects analysis followed with multiple comparisons test. **p < 0.01. See Source Data Extended Data Fig. 6 for detailed values and statistics.
Extended Data Fig. 7 Histology and light-induced c-Fos in the MPOAEsr1 and BNSTprEsr1 terminal manipulation experiments.
(a) Experimental design to optogenetically inactivate MPOAEsr1 inputs to BNSTpr. (b) Representative histology of 6 mice showing ArchT expression (red) in the MPOA (left), and ArchT expression terminals and optic fibre tracks in the BNSTpr (right). (c) Experimental design to optogenetically activate MPOAEsr1 inputs to the BNSTpr. (d) Representative histology of 7 mice showing ChrimsonR (red) and c-Fos (white) in the MPOA after delivering 5 mW pulsed yellow light to the right side of the BNSTpr for 10 min. (e) Representative histology of 3 mice showing ChR2 (green), hM4Di (red), and c-Fos (white) in the MPOA after delivering 10 min 5 mW pulsed blue light to the right side of the BNSTpr 30 min after CNO injection. (f) Number of c-Fos+ cells in the right MPOA in ChrimsonR and ChR2+Gi group. Error bars: ± SEM. Unpaired t test, ****p < 0.0001. n = 7 mice for ChrimsonR group, n = 3 for ChR2+Gi group. (g) Experimental design to optogenetically inactivate BNSTprEsr1 inputs to MPOA. (h) Representative histology of 6 mice showing ArchT (red) in the BNSTpr (left), and ArchT expressing axons and optic fibre tracks in the MPOA (right). (i) Experimental design to optogenetically activate BNSTprEsr1 inputs to MPOA. (j) Representative histology of 5 mice showing ChrimsonR (red) and c-Fos (white) in the BNSTpr after delivering 5 mW pulsed yellow light to the right side of the MPOA for 10 min. (k) Representative histology of 4 mice showing ChR2 (green), hM4Di (red) and c-Fos (white) in BNSTpr after delivering 10 min 5 mW pulsed blue light to the right MPOA 30 min after CNO injection. (l) Number of c-Fos+ cells in the right BNSTpr in ChrimsonR and ChR2+Gi group. Error bars: ± SEM. Mann Whitney test, *p < 0.05. n = 5 mice for ChrimsonR group, n = 4 for ChR2+Gi group. See Source Data Extended Data Fig. 7 for detailed values and statistics.
Extended Data Fig. 8 Verification of ArchT-mediated terminal inhibition.
(a and d) Experimental design to examine the efficacy of ArchT-mediated inhibition of MPOAEsr1 → BNSTprEsr1 (a) and BNSTprEsr1 → MPOAEsr1 (d) projections. (b and e) Representative blue light pulse evoked IPSCs from BNSTprEsr1 (b) and MPOAEsr1cells (e) with (yellow) and without (black) simultaneous 5 mW yellow light delivery. (c and f) The amplitude of oIPSCs of BNSTprEsr1 (c) and MPOAEsr1 cells (f) with and without 5 mW yellow light delivery. Error bars: ± SEM. Paired t-test (c) and Wilcoxon matched-pairs signed rank test (f), **p < 0.01. n = 4 BNSTprEsr1 cells, n = 8 MPOAEsr1 cells. See Source Data Extended Data Fig. 8 for detailed values and statistics.
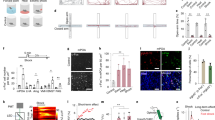
Extended Data Fig. 9 Fibre photometry recording of GFP-expressing BNSTprEsr1 and MPOAEsr1 cells and GCaMP6f-expressing BNSTprMPOA cells during pup interaction.
(a) Fibre photometry setup. (b and d) Viral construct and targeted brain regions. Brain illustrations are produced based on reference atlas from https://atlas.brain-map.org/. (c and e) Representative histological images of 4 mice showing GFP (green) expression and fibre tracks (white box) in BNSTpr (c) and MPOA (e). (f) Experimental timeline. (g,k) Representative GFP recording (ΔF/F) traces of BNSTprEsr1 (g) and MPOAEsr1 (k) cells during pup interaction in a hostile SW virgin female (g1, k1) and a mother (g2, k2). Colour shades indicate various behaviours. (h,l) PETHs of GFP signal (Z-scored ΔF/F) of BNSTprEsr1 (h) and MPOAEsr1 (l) cells aligned to the first pup contact in hostile virgin females (h1, l1), and mothers (h2, l2). n = 4 mice. Solid lines indicate the onset on first pup contact; dashed black lines indicate the mean duration of first pup contact. Shades: ± SEM. (i,m) PETHs of GFP signal (Z-scored ΔF/F) of BNSTprEsr1 (i) and MPOAEsr1 (m) cells aligned to the onset of pup approach in hostile virgin females (i1, m1), and mothers (i2, m2). n = 4 mice for each group. Blue dashed lines indicate the onset of pup approach; green dashed lines indicate the mean latency to pup investigation; the red and black dashed lines in i1 and m1 indicate the mean latency to attack and stop attacking pups, respectively; the magenta and black dashed lines in i2 and m2 indicate the mean latency to retrieve and stop retrieving pups, respectively. Shades: ± SEM. (j,n) Mean AUC of Z-scored ΔF/F signal of BNSTprEsr1 (j) and MPOAEsr1 (n) cells during various pup-directed behaviours in hostile virgin females and mothers. Two-way RM ANOVA (j) and Mixed-effects analysis (n) followed with multiple comparisons test. n = 4 mice for each group. Error bars: ± SEM. (o) Experimental design. (p) A representative histological image of 9 mice showing GCaMP6f (green) expression and the fibre track in BNSTpr (white lines). (q) Experimental timeline. (r) Representative GCaMP6f recording (ΔF/F) traces of BNSTprMPOA cells during pup interaction in a hostile SW virgin female (r1) and a mother (r2). Colour shades indicate various behaviours. (s–v) PETHs of GCaMP6f signal (Z-scored ΔF/F) of BNSTprMPOA cells aligned to the onset of various behaviours in hostile virgin females (s1-v1), and mothers (s2-v2). n = 6 mice. Solid lines indicate the onset on each behaviour; dashed black lines indicate the end of each behaviour. Shades: ± SEM. (w) Mean AUC of Z-scored ΔF/F signal of BNSTprMPOA cells during the first pup contact. Paired t-test. **p < 0.01. Error bars: SEM. n = 6 mice. (x,y) Mean AUC of Z-scored ΔF/F signal of BNSTprMPOA cells during pre-pup period and various pup-directed behaviours in hostile virgin females and mothers. Two-way RM ANOVA followed with multiple comparisons test. **p < 0.01, ****p < 0.0001, mean ± SEM. n = 6 mice. See Source Data Extended Data Fig. 9 for detailed values and statistics.
Extended Data Fig. 10 Simultaneous recording of MPOAEsr1 and BNSTprEsr1 cells and comparing infanticide and maternal behaviour-induced c-Fos between strains.
(a) Fibre photometry setup. (b) Viral construct and targeted brain regions. Brain illustrations are produced based on reference atlas from https://atlas.brain-map.org/. (c,d) Representative histological images of 3 mice showing GCaMP6f (green) expression and fibre tracks (white boxes) in MPOA (c) and BNSTpr (d). (e) Experimental timeline. (f) Representative GCaMP6f recording (ΔF/F) traces of MPOAEsr1 (black) and BNSTprEsr1 (purple) cells during pup interaction in a hostile SW virgin female (f1) and a mother (f2). Colour shades indicate various behaviours. (g) PETHs of GCaMP6f signal (Z-score ΔF/F) of MPOAEsr1 (black) and BNSTprEsr1 (purple) cells aligned to the onset of pup approach in hostile virgin females (g1), and mothers (g2). n = 3 mice for each group. Blue dashed lines indicate the onset of pup approach; green dashed lines indicate the mean latency to pup investigation; the red and black dashed lines in g1 indicate the mean latency to attack and stop attacking pups, respectively; the magenta and black dashed lines in g2 indicate the mean latency to retrieve and stop retrieving pups, respectively. Shades: ± SEM. (h) Mean AUC of Z-scored ΔF/F signal of MPOAEsr1 (black) and BNSTprEsr1 (purple) cells during pup approach, investigation, attack, and retrieval in hostile virgin females (h1) and mothers (h2). Two-way RM ANOVA (h1) and Mixed-effects analysis (h2) followed with multiple comparisons test. n = 3 mice for each group, *p < 0.05, ***p < 0.001, mean ± SEM. (i) BNSTprEsr1 and MPOAEsr1 response ratio in hostile virgin females and mothers during pup interaction period. The red dashed line indicates 1. n = 3 mice for each group, Paired t-test. *p < 0.05, mean ± SEM. (j) Representative histological images of 3 mice in each group showing c-Fos expression in MPOA and BNSTpr of a C57BL/6 virgin maternal female (left), a SW virgin maternal female (middle), and a SW infanticidal female (right). (k, l) Number of c-Fos+ cells in MPOA (k) and BNSTpr (l) in C57BL/6 virgin maternal, SW virgin maternal and SW infanticidal female mice. n = 3 for each group, ***p < 0.001, ****p < 0.0001. Error bars: ± SEM. One-way ANOVA followed with multiple comparisons test. See Source Data Extended Data Fig. 10 for detailed values and statistics.
Supplementary information
Supplementary Notes
Supplementary Notes 1 and 2.
Supplementary Data
Source data for Supplementary Note 1
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mei, L., Yan, R., Yin, L. et al. Antagonistic circuits mediating infanticide and maternal care in female mice. Nature 618, 1006–1016 (2023). https://doi.org/10.1038/s41586-023-06147-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06147-9
This article is cited by
-
Independent inhibitory control mechanisms for aggressive motivation and action
Nature Neuroscience (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.