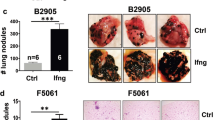
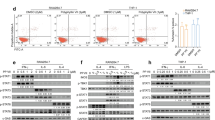
Abstract
Metastasis frequently develops from disseminated cancer cells that remain dormant after the apparently successful treatment of a primary tumour. These cells fluctuate between an immune-evasive quiescent state and a proliferative state liable to immune-mediated elimination1,2,3,4,5,6. Little is known about the clearing of reawakened metastatic cells and how this process could be therapeutically activated to eliminate residual disease in patients. Here we use models of indolent lung adenocarcinoma metastasis to identify cancer cell-intrinsic determinants of immune reactivity during exit from dormancy. Genetic screens of tumour-intrinsic immune regulators identified the stimulator of interferon genes (STING) pathway as a suppressor of metastatic outbreak. STING activity increases in metastatic progenitors that re-enter the cell cycle and is dampened by hypermethylation of the STING promoter and enhancer in breakthrough metastases or by chromatin repression in cells re-entering dormancy in response to TGFβ. STING expression in cancer cells derived from spontaneous metastases suppresses their outgrowth. Systemic treatment of mice with STING agonists eliminates dormant metastasis and prevents spontaneous outbreaks in a T cell- and natural killer cell-dependent manner—these effects require cancer cell STING function. Thus, STING provides a checkpoint against the progression of dormant metastasis and a therapeutically actionable strategy for the prevention of disease relapse.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw sequencing data corresponding to CRISPR screens and targeted bisulfite sequencing have been deposited in the NCBI Sequence Read Archive under accessions PRJNA786579 and PRJNA867723, respectively. ChIP–seq data have been deposited in the Gene Expression Omnibus under accession GSE210946. Source data are provided with this paper.
Code availability
Custom code used in this study are available at Zenodo: https://doi.org/10.5281/zenodo.7618821.
References
Goddard, E. T., Bozic, I., Riddell, S. R. & Ghajar, C. M. Dormant tumour cells, their niches and the influence of immunity. Nat. Cell Biol. 20, 1240–1249 (2018).
Malladi, S. et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 165, 45–60 (2016).
Pommier, A. et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 360, eaao4908 (2018).
Pantel, K. et al. Frequent down-regulation of major histocompatibility class I antigen expression on individual micrometastatic carcinoma cells. Cancer Res. 51, 4712–4715 (1991).
Eyles, J. et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J. Clin. Invest. 120, 2030–2039 (2010).
Massagué, J. & Ganesh, K. Metastasis-initiating cells and ecosystems. Cancer Discov. 11, 971–994 (2021).
Consonni, D. et al. Lung cancer prognosis before and after recurrence in a population-based setting. JNCI 107, djv059 (2015).
Willis, R. A. The Spread of Tumours in the Human Body (J. & A. Churchill, 1934).
Hadfield, G. The dormant cancer cell. Br. Med. J. 2, 607–610 (1954).
Janni, W. et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse—a European pooled analysis. Clin. Cancer Res. 17, 2967–2976 (2011).
Sosa, M. S., Bragado, P. & Aguirre-Ghiso, J. A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14, 611–622 (2014).
Xiao, D. et al. Donor cancer transmission in kidney transplantation: a systematic review. Am. J. Transplant. 13, 2645–2652 (2013).
Laughney, A. M. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 26, 259–269 (2020).
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011).
Marcus, A. et al. Recognition of tumors by the innate immune system and natural killer cells. Adv. Immunol. 122, 91–128 (2014).
McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science 363, eaat8657 (2019).
Chen, Q. et al. Carcinoma–astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018).
Hong, C. et al. cGAS–STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 (2022).
Konno, H., Konno, K. & Barber, G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Chen, H. et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147, 436–446 (2011).
Nguyen, D. X. et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138, 51–62 (2009).
Luis-Ravelo, D. et al. Tumor–stromal interactions of the bone microenvironment: in vitro findings and potential in vivo relevance in metastatic lung cancer models. Clin. Exp. Metastasis 28, 779–791 (2011).
Jeremiah, N. et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124, 5516–5520 (2014).
Abe, T. et al. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 50, 5–15 (2013).
Dufour, J. H. et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168, 3195–3204 (2002).
Appay, V. & Rowland-Jones, S. L. RANTES: a versatile and controversial chemokine. Trends Immunol. 22, 83–87 (2001).
Schutyser, E., Struyf, S. & Van Damme, J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14, 409–426 (2003).
Kitapma, S. et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov. 9, 34–45 (2019).
Bragado, P. et al. TGF-β2 dictates disseminated tumour cell fate in target organs through TGF-β-RIII and p38α/β signalling. Nat. Cell Biol. 15, 1351–1361 (2013).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
David, C. J. & Massagué, J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018).
Su, J. et al. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577, 566–571 (2020).
Pan, B. S. et al. An orally available non-nucleotide STING agonist with antitumor activity. Science 369, eaba6098 (2020).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Flood, B. A., Higgs, E. F., Li, S. Y., Luke, J. J. & Gajewski, T. F. STING pathway agonism as a cancer therapeutic. Immunol. Rev. 290, 24–38 (2019).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Chen, J. et al. Cell cycle checkpoints cooperate to suppress DNA- and RNA-associated molecular pattern recognition and anti-tumor immune responses. Cell Rep. 32, 108080 (2020).
Deng, L. F. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).
Woo, S. R. et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Zierhut, C. et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e323 (2019).
Ahn, J. et al. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014).
Vasudevan, A. et al. Single-chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev. Cell 52, 413–428.e6 (2020).
Tello-Lafoz, M. et al. Cytotoxic lymphocytes target characteristic biophysical vulnerabilities in cancer. Immunity 54, 1037–1054.e1037 (2021).
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009).
Girardin, S. E. et al. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300, 1584–1587 (2003).
Wang, Q. et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41, 919–933 (2014).
Soto-Feliciano, Y. M. et al. A molecular switch between mammalian MLL complexes dictates response to Menin–MLL inhibition. Cancer Discov. 13, 146–169 (2022).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Michlits, G. et al. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat. Methods 17, 708–716 (2020).
Sánchez-Rivera, F. J. et al. A base editing sensor streamlines high-throughput guide validation and engineering of cancer associated variants. Nat. Biotechnol. 40, 862–873 (2022).
Dow, L. E. et al. Inducible in vivo genome editing with CRISPR–Cas9. Nat. Biotechnol. 33, 390–394 (2015).
Minn, A. J. et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest. 115, 44–55 (2005).
Ebright, R. Y. et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 (2020).
Oshimori, N., Oristian, D. & Fuchs, E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015).
Ahn, J., Gutman, D., Saijo, S. & Barber, G. N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl Acad. Sci. USA 109, 19386–19391 (2012).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Neri, S., Mariani, E., Meneghetti, A., Cattini, L. & Facchini, A. Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab. Immunol. 8, 1131–1135 (2001).
Chava, S., Bugide, S., Gupta, R. & Wajapeyee, N. Measurement of natural killer cell-mediated cytotoxicity and migration in the context of hepatic tumor cells. J. Vis. Exp. https://doi.org/10.3791/60714 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Karpova, N. N. & Umemori, J. Protocol for methylated DNA immunoprecipitation (MeDIP) analysis. Epigenetic Methods Neurosci. Res. 105, 97–114 (2016).
Acknowledgements
The authors thank the Thoracic Oncology Service, the Pathology Core Facility, the Flow Cytometry Core, the Molecular Cytology Core, the Genomic Editing and Screening Core, the Single-cell Analysis Innovation Lab, the Integrated Genomics Operation, and the Research Animal Resource Center at MSKCC for their assistance; the tissue donors at MSKCC for participating in this study; E. Er for early guidance with metastasis assays; C. Rudin for assistance in the procurement of tumour tissue samples; and Z. Zhang, H.-A. Chen and V. Tem for technical assistance. This work was supported by NIH grants R35-CA252978 and P01-CA129243 (J.M.), U54-CA209975 (J.M. and D.P.), K08-CA230213 (K.G.), P30 CA008748 (MSKCC), the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center at MSKCC (J.H., F.J.S.-R., Z.W., K.G., D.P. and J.M.), the Agilent Technologies Thought Leader Award (S.W.L.), and postdoctoral fellowships from the Terri Brodeur Breast Cancer Foundation (J.H.), the Translational Research Oncology Training Program 5T32CA160001 (F.J.S.-R.), and the Damon Runyon Quantitative Biology Program (S.G.). F.J.S.-R. is a HHMI Hanna Gray Fellow. D.P. and S.W.L. are HHMI Investigators.
Author information
Authors and Affiliations
Contributions
J.H. and J.M. conceived studies, designed experiments, interpreted results and wrote the manuscript. J.H. performed experiments and analysed data. F.J.S.-R., Y.-j.H. and S.W.L. designed and analysed CRISPR screens. S.G., S.K. and D.P. performed scRNA-seq data analysis. G.N.J. and J.P.H. provided technical assistance with experiments. Z.W. generated and characterized TGFβ-reporter and TGFBR2-knockout cell lines and performed immunohistochemistry analysis. K.G. established the KPad1 cell line. K.G., S.U. and C.A.I.-D. procured patient-derived tumour samples and validated the STING immunohistochemistry scoring. A.M.M., R.B.D. and J.C.S. performed immune phenotyping and NK cell cytotoxicity experiments. H.Z. and E.d.S. conducted drug treatment experiments in mice.
Corresponding author
Ethics declarations
Competing interests
S.W.L. received funding and research support from Agilent Technologies for the purposes of massively parallel oligonucleotide synthesis to generate the sgRNA libraries described in this Article.
Peer review
Peer review information
Nature thanks Melanie Werner-Klein and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 In vivo CRISPR screen for cell-autonomous immune regulators of dormant metastasis.
(a) Metastasis-free survival Kaplan-Meier plots of athymic mice intracardially inoculated with 1 x 105 H2030-BrM or H2087-LCC cells. One set of mice inoculated with H2087-LCC cells was treated with anti-asialo-GM1 antibody to deplete NK cells 30 days after cell inoculation. n = 9 (H2030-BrM) or 10 (H2087-LCC) mice per group, log-rank test. (b) Flow cytometry analysis of Lineage–CD45+NKP46+ NK cells in blood from athymic mice that were treated with IgG control or anti-asialo-GM1 antibody for 4 days. Lineage– : TCRβ–CD3–CD19–B220–CD11c–Ly6G–F4/80–. n = 3 mice per group. Mean ± s.e.m., two-sided unpaired t-test. (c) Petal charts of metastasis incidence in specific organs of 10 athymic mice intracardially inoculated with 1x105 H2087-LCC cells, treated with or without anti-asialo-GM1 antibody. Metastasis progression was monitored by BLI for 160 days after cell inoculation. (d) Petal charts of metastasis incidence in specific organs after intracardiac inoculation of 2x104 KPad1 cells into 8-9 B6-albino mice or NSG mice. Metastasis progression was monitored by BLI for 60 days (B6-albino mice) or 28 days (NSG mice) after cell inoculation. (e) Flow cytometry analysis of Lineage–CD45+NKP46+ NK cells, CD45+CD3+CD4+, and CD45+CD3+CD8+ T cells in blood from B6-albino mice that were treated for 3 days with IgG control, anti-NK1.1, anti-CD4, or anti-CD8 antibodies. n = 3 mice per group. Mean ± s.e.m., two-sided unpaired t-test. (f) Metastasis-free survival Kaplan-Meier plots of NSG mice inoculated intracardially with KPad1 cells, or B6-albino mice inoculated intracardially with KPad1 cells or KP-482T1 cells, and treated with the indicated antibodies to deplete specific types of immune cells. 1 x 105 cells were inoculated per mouse. Antibody treatments started 12 days after inoculation. n = 8 (KPad1/control, KPad1/αCD4), 7 (KPad1/αNK1.1, KPad1/αCD8, KP-482T1), or 9 (NSG) mice per group, log-rank test. *, p = 0.03 (KPad1/αNK1.1 vs. KPad1/control), *, p = 0.0146 (KPad1/αCD8 vs. KPad1/control), **, p = 0.0094 (KPad1/αCD4 vs. KPad1/control), ***, p = 0.0002 (KP-482T1 vs. KPad1/control), ****, p < 0.0001 (KPad1/NSG vs. KPad1/control in B6-albino). (g) Gene set enrichment analysis (GSEA) showing pathways enriched in dormant H2087-LCC cells that were induced to proliferate in culture compared to cells from spontaneous metastatic outbreaks, analyzed from previous scRNA-seq data sets. (h–i) Metastasis-free survival of athymic mice (h) or B6-albino mice (i) intracardially inoculated with 2.5 x 105 H2087-LCC (h) or KPad1 cells (i), expressing a scrambled RNA control or various sgRNA pools. n = 9 (Scrambled), 10 (NK ligands and MHC class I), 12 (STING pathway), 10 (LPS and RNA-sensing pathways), or 11 (Interferon response, Complement pathway) mice per group (h); n = 7 (Scrambled, LPS and RNA-sensing pathways), 8 (Complement pathway), or 9 (other groups) mice per group (i). (j) BLI quantification of lung metastatic colonies after intravenous inoculation of 2.5 x 105 KPad1 cells expressing a scrambled control or different sgRNA pools in B6-albino mice. Tissues above the dotted line were harvested 57 days after cell inoculation, then subjected to sgRNA recovery and analysis. n = 9 (Additional MHC class I) or 10 (other groups) mice per group. (k–l) Gene rank based on the average fold enrichment of sgRNAs in multi-organ metastases from H2087-LCC (k) or KPad1 (l) cells. (m) Gene rank based on average fold change of sgRNAs enriched in the lung colonies formed after tail-vein inoculation of 2.5 x 105 KPad1 cells into B6-albino mice. For k-m, genes with average log2FC >0 are plotted and genes targeted by the top 10 enriched sgRNAs are listed.
Extended Data Fig. 2 sgRNAs enriched in metastases of specific organs.
(a–b) Dot plots showing genes targeted by the enriched sgRNAs in metastases of the indicated organs after intracardiac inoculation of 2.5 x 105 H2087-LCC cells into athymic mice (a) or 2.5 x 105 KPad1 cells into B6-albino mice (b). Genes with average log2FC >0 are plotted.
Extended Data Fig. 3 STING in dormant and reactivated metastatic progenitors and progenies.
(a) As in Fig. 2a, representative IF images of single quiescent (Ki67low) and proliferative (Ki67high) cells during dormancy, and spontaneous outbreaks (SO) from H2087-LCC, stained for CCL5 IF (red), human vimentin (green), Ki67 IF (white), and DAPI (blue). The staining signal intensity of CCL5 in vimentin-positive cells was quantified and plotted (a.u., arbitrary units). Scale bar: 10 μm. n = 89 cells (Ki67low), 24 cells (Ki67high), or 45 regions (outbreaks), from 5 mice per group. Mean ± s.e.m, two-sided unpaired t-test. (b–c) Representative IF staining images of KPad1 cells (GFP, green) in brains of B6-albino mice that were intracardially inoculated with 1 x 105 KPad1 cells. Organs were harvested 25 days after inoculation to capture the dormant state, and 43–61 days after inoculation to capture spontaneous outbreaks. Tissue sections were stained for STING (b, red), CCL5 (c, red), Ki67 (b and c, white), and DAPI (b and c, blue), and the staining signal intensity of STING (b) and CCL5 (c) in GFP-positive cells was quantified and plotted (a.u., arbitrary units). Scale bar: 20 μm. n = 16 cells (Ki67low), 7 cells (Ki67high), or 22 lesions (outbreaks), from 4 mice per group in (b); n = 15 cells (Ki67low), 10 cells (Ki67high), or 33 lesions (outbreaks), from 4 mice per group in (c). Mean ± s.e.m, two-sided unpaired t-test. (d) Representative images of micrometastatic cells (cytokeratin AE1/AE3, green) in lymph nodes from stage II and stage III LUAD patients, stained for CCL5 IF (red), Ki67 (white), and DAPI (blue). The signal intensity of CCL5 in cytokeratin-positive cells was quantified and plotted. Scale bar: 20 μm. n = 24 (Ki67low) or 44 (Ki67high) regions, from 14 lymph nodes of 9 patients. Mean ± s.e.m., two-sided unpaired t-test. (e) Representative images of STING IHC intensity levels (0, none; 1, weak; 2, moderate; 3, strong) in patient-derived lung adenocarcinoma tissue samples. STING expression H-scores were calculated based on the intensity levels, representative of 2 independent experiments. Scale bar: 50 μm. (f–g) IF staining (f) and quantification (g) of CCL5 expression in patient-derived LUAD metastases compared to matched primary tumors. Tissue sections were stained for cytokeratin AE1/AE3 IF (green), CCL5 IF (red), and DAPI (blue), and the staining signal intensity of CCL5 in cytokeratin-positive areas was quantified and plotted (a.u., arbitrary units). Scale bar: 50 μm. n = 4 matched pairs, two-tailed paired t-test. (h) Expression of SOX transcription factors specifying early (SOX2) and late (SOX9) lung epithelial progenitor states, STING, canonical STING pathway targets, hallmark_interferon α response genes (GSEA), canonical and non-canonical NF-kB target genes (rows) across H2087-LCC single cells isolated from dormant metastases or spontaneous macrometastases in athymic mice, and cultured under growth-promoting conditions. STING pathway targets refers to the mean expression of the genes shown in Extended Data Fig. 4. For each gene, imputed expression was z-normalized across all cells. In the top row, cells (columns) are colored and labeled by source. n = 2245 cells isolated from 3 mice. (i) Violin plots showing imputed and z-normalized expression of STING in H2087-LCC cells from the data set used in (h). n = 2245 cells isolated from 3 mice. Two-sided Mann-Whitney U-test. (j) Expression of SOX transcription factors, STING, canonical STING pathway targets, hallmark interferon α response genes (GSEA), canonical and non-canonical NF-kB target genes (rows) across patient-derived metastatic tumor cells assigned to early or late-stage progenitor states. STING pathway targets refers to the mean expression of the genes shown in Extended Data Fig. 4. For each gene, imputed expression was z-normalized across all cells and smoothed using a 20-cell moving average window. Individual cells (columns) are ranked left to right by average lung epithelial development score. n = 991 cells isolated from 5 patients. (k) Violin plots showing imputed and z-normalized expression of STING across patient-derived tumor cells assigned to early or late-stage progenitor states from the data set used in (j). n = 991 cells isolated from 5 patients. Two-sided Mann-Whitney U-test. (l) Clustered heatmap showing expression of SOX2, SOX9, and STING in H2087-LCC cells isolated from metastases formed after NK cell depletion by anti-asialo-GM1 antibody treatment in athymic mice. For each gene, imputed expression was z-normalized across all cells. n = 6073 cells isolated from 5 mice. (m) qRT-PCR analysis of STING mRNA levels in H2087-LCC cell cultures derived from spontaneous outbreaks (SO) compared to those derived from outbreaks following depletion of NK cells. NK cells were depleted by treating mice with anti-asialo-GM1 antibody 30 days after cancer cell inoculation. n = 9 H2087-LCC cell lines from 4 mice after NK cell depletion or n = 17 cell lines from 8 mice with spontaneous outbreaks. Mean ± s.e.m., two-sided unpaired t-test. (n) IHC staining and quantification of STING expression (H-score) in H2087-LCC spontaneous outbreaks and outbreaks following NK cell depletion in multiple organs from athymic mice. Scale bar: 50 μm. n = 3 organs from 3 mice (NK depletion) or 7 organs from 3 mice (spontaneous). Mean ± s.e.m., two-sided unpaired t-test.
Extended Data Fig. 4 Expression of canonical STING pathway targets from scRNA-seq datasets.
(a) Expression of SOX transcription factors specifying early (SOX2) and late (SOX9) lung epithelial progenitor states, STING and canonical STING pathway targets (rows) across H2087-LCC single cells isolated from dormant metastases, or spontaneous macrometastases in athymic nude mice, and cultured under growth-promoting conditions. STING pathway targets refers to the mean expression of the genes shown in the bottom heatmap panel. For each gene, imputed expression was z-normalized across all cells. In the top row, cells (columns) are colored and labeled by source. n = 2245 cells isolated from 3 mice. (b) Expression of SOX transcription factors, STING and canonical STING pathway targets (rows) across patient-derived metastatic tumor cells assigned to early or late-stage progenitor states. STING pathway targets refers to the mean expression of the genes shown in the bottom heatmap panel. For each gene, imputed expression was z-normalized across all cells and smoothed using a 20-cell moving average window. Individual cells (columns) are ranked left to right by average lung epithelial development score, as in Laughney et al. n = 991 cells isolated from 5 patients.
Extended Data Fig. 5 Perturbation of STING activity in human LUAD models.
(a) Western immunoblot analysis of STING and β-actin in WT and STING knockout H2087-LCC cells, representative of 2 independent experiments. (b–c) Immunoblotting (b) and qRT-PCR analyses (c) of STING expression in parental H2087-LCC cells and the spontaneous outbreak derivatives SO1 and SO2. Mean ± s.e.m, representative of 2 independent experiments. In (c), each dot represents a technical replicate of the assay. (d) qRT-PCR analysis of IFNB1 expression after cGAMP treatment of H2087-LCC, SO1, and SO2 cells. Mean ± s.e.m, representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (e) Western immunoblot analysis of STING (endogenous STING and HA-tagged STING) and β-actin in H2087-LCC-SO cells inducibly expressing STING-HA under doxycycline (Dox) treatment for 24 h. Representative of 2 independent experiments. (f) Metastasis-free survival plots of athymic mice inoculated intracardially with 1 x 105 H2087-LCC-SO cells. Cells were transduced with a vector control or a Dox-inducible STING expression vector. Dox treatment started 7 days after cell inoculation. n = 13 (Control, STING), 10 (Control+Dox), 11 (STING+Dox) mice per group, log-rank test. (g) qRT-PCR analysis of STING mRNA levels in human primary bronchial epithelial cells, H2087, A549, and H2030 LUAD cell lines. Mean ± s.e.m, representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (h–i) qRT-PCR analysis of IFNB1 mRNA expression in human primary bronchial epithelial cells and lung adenocarcinoma cell lines treated with cGAMP (h) or poly(I:C) (i). Mean ± s.e.m, representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (j) BLI and quantification of metastases formed by H2030-BrM cells inducibly expressing activated STING-V155M, starting the day after intracardiac inoculation of 1 x 105 cells in athymic mice. Metastases were quantified 21 days after inoculation. n = 9 mice per group. Mean ± s.e.m., two-sided Mann-Whitney U-test. For gel source data, see Supplemental Fig. 1.
Extended Data Fig. 6 STING activity in mouse lung adenocarcinoma cells.
(a-b) qRT-PCR analysis of Sting (a) and Cxcl10 (b) mRNA levels after cGAMP treatment in KPad1 and KP-482T1 cells. Mean ± s.e.m, representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (c) Western immunoblotting of STING and β-actin in parental and Sting knockout KPad1 cells. Representative of 2 independent experiments. For gel source data, see Supplemental Fig. 1. (d) RFP-positive WT KPad1 cells and GFP-positive Sting KO KPad1 cells were mixed at the indicated ratios and intracardially inoculated (1 x 105 cells) in B6-albino mice. Metastases-bearing organs were harvested 4-17 weeks after inoculation and the percentage of GFP-positive cells and RFP-positive cells was determined by flow cytometry. n = 9 (1:1, 9:1) or 5 (3:1) lesions from 3-5 mice per group. Mean ± s.e.m. (e) Percentage of EdU positive cells of WT or Sting knockout KPad1 cells incubated with EdU for 2 h. n = 3 per group. Mean ± s.e.m. (f) Percentage of EdU positive cells in KP-482T1 Tet-On Sting cultures treated with Dox for 24 h and then incubated with EdU for 2 h. n = 3 per group. Mean ± s.e.m. (g–h) BLI signal intensity at the indicated time points after intracardiac inoculation of 1 x 105 WT or Sting knockout KPad1 cells (g) or KP-482T1 cells with induced overexpression of STING (h) in B6-albino mice. n = 16 (WT) or 15 (KO) mice (g); n = 7 (control) or 6 (Dox) mice (h). Mean ± s.e.m. (i) Heatmap representation of cytokine array analysis of culture supernatant from KP-482T1 cells transduced with a vector control or a Sting overexpression vector. n = 3 for each condition.
Extended Data Fig. 7 STING knockout and overexpression effects on growth and differentiation of metastatic progenitors.
(a) Percentage of EdU positive cells of WT or STING knockout H2087-LCC cells incubated with EdU for 2 h. n = 3 per group. Mean ± s.e.m. (b) Percentage of EdU positive cells in H2087-LCC-SO Tet-On STING or H2030-BrM Tet-On STINGV155M cultures treated with Dox for 24 h, and then incubated with EdU for 2 h. n = 3 per group. Mean ± s.e.m. (c-d) BLI signal intensity at the indicated time points after intracardiac inoculation of 1 x 105 WT or STING knockout H2087-LCC cells (c) or H2030-BrM cells with Dox-induced overexpression of STINGV155M (d) in athymic mice. n = 7 (WT, KO1) or 6 (KO2) mice per group (c); n = 9 (7 days) or 7 (other groups) mice per group (d). Mean ± s.e.m. (e) IF staining analysis showing percentage of SOX2high and SOX9high cells in spontaneous metastases formed after intracardiac inoculation of 1 x 105 WT or STING knockout (KO) H2087-LCC cells in athymic mice. Organs were harvested 12 weeks after inoculation. n = 10 lesions (WT) or 23 lesions (STING KO). Mean ± s.e.m. (f) IF staining analysis showing percentage of SOX2high and SOX9high cells in metastases formed after intracardiac inoculation of 1 x 105 H2087-LCC-SO cells inducibly overexpressing STING in athymic mice. Organs were harvested 7–13 weeks after inoculation for the control cells and 18 weeks after inoculation for the STING overexpressing cells. n = 48 images from 5 mice (control) or 13 images from 3 mice (Dox). Mean ± s.e.m.
Extended Data Fig. 8 Cancer cell STING increases NK and T cell levels in metastases.
(a) Schematic of flow cytometry or immunofluorescence analysis of immune cells in bone metastasis. (b) Flow cytometry analysis and quantification of CD45+ leukocytes, CD45+CD19−TCRβ−XCR1+CD11c+ cDC1 cells, CD45+NK1.1+TCRβ−CD49b+NKp46+ NK cells, CD45+TCRβ+NK1.1−CD8+ T cells, and CD45+TCRβ+NK1.1−CD4+ T cells in KP-482T1 metastasis-bearing femurs after Dox-induced expression of STING for 2 days or 7 days. Cell numbers per gram of femur tissue were counted and normalized to control (-Dox). Metastasis-bearing femurs were collected 2 weeks after intracardiac inoculation of 2 x 104 KP-482T1 cells in B6-albino mice. n = 7 mice per group. Mean ± s.e.m., two-sided unpaired t-test. p values comparing each Dox treatment group with control (−Dox). (c–f) Percentage of NK cells degranulating (CD107a+) (c) or producing IFNγ (d), and percentage of CD44+CD8+ T cells producing IFNγ (e) or TNF (f), isolated from KP-482T1 metastasis-bearing femurs after Dox-induced expression of STING for 2 days or 7 days, and then cultured ex vivo with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 4h. n = 5 (+Dox 7 days) or 6 (other groups) mice per group. Mean ± s.e.m. (g) Schematic of the NK cell-mediated killing assay, and the percentage of WT or STING KO H2087-LCC cells killed by incubation with naïve NK cells for 4h at the indicated effector:target ratios. n = 3 per group. Mean ± s.e.m. (h) Schematic of the trans-well migration assay, and the number of NK cells migrated into cell culture media conditioned by WT or STING knockout H2087-LCC cells. n = 3 per group. Mean ± s.e.m., two-sided unpaired t-test.
Extended Data Fig. 9 STING locus regulation during metastatic progression.
(a) Gene track view for H3K27ac, H3K4me1, and H3K4me3 ChIP-Seq tags at the STING locus in H2087-LCC, spontaneous outbreak derivative H2087-LCC-SO1, and H2030 cells. (b) ChIP-PCR analysis of DNMT3B binding to the STING promoter or 3′ enhancer regions in parental H2087-LCC, H2087-LCC-SO1, H2087-LCC-SO2, and in A549 and H2030 cells. Mean ± s.e.m., representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (c) qRT-PCR analysis of STING mRNA levels in H2030 and A549 cells after a 3-day treatment with 100nM 5-aza-2′deoxycytidine (5-azadC) in culture. Mean ± s.e.m., representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (d) Schematic of targeted bisulfite sequencing analysis of the STING locus in metastatic cells of interest.
Extended Data Fig. 10 TGF-β mediated suppression of STING expression.
(a) Growth rate of H2087-LCC cells treated with TGF-β or CDK4/6 inhibitor palbociclib for the indicated times. n = 4. Mean ± s.e.m., two-way ANOVA analysis. (b) qRT-PCR analysis of STING mRNA levels in H2087-LCC cells treated with TGF-β or palbociclib for 7 days. Mean ± s.e.m., representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (c) qRT-PCR analysis of IFNB1 and CCL5 expression in WT or STING knockout H2087-LCC cells treated with TGF-β for 3 days. Mean ± s.e.m., representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (d) qRT-PCR analysis of cGAS mRNA levels in H2087-LCC cells treated with TGF-β for 7 days. Mean ± s.e.m., representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (e) ELISA analysis of cGAMP levels in H2087-LCC cells treated with TGF-β for 5 days. n = 3 per group. Mean ± s.e.m. (f) Gene track view of H3K27ac, H3K4me1, and H3K4me3 ChIP-Seq tags at the STING locus in H2087-LCC cells treated with TGF-β for 4 days. (g) PCR based quantitation of methylated STING promoter and 3′ enhancer sequences in MeDIP samples from H2087-LCC cells that were treated with TGF-β for the indicated times. Mean ± s.e.m., representative of 3 independent experiments. Each dot represents a technical replicate of the assay. (h) Schematic of the TGF-β and SMAD-dependent transcriptional reporter. SBE: Smad Binding Element. mCMV: minimal CMV promoter. TRE: Tetracycline-Responsive promoter Element. (i) Representative IF staining images of H2087-LCC cells (human vimentin, green) expressing SMAD-responsive mCherry reporter in the lungs of athymic mice intravenously inoculated with 1x105 cells. Organs were harvested 8 weeks after inoculation. Disseminated cancer cells were present as single quiescent (Ki67low) or proliferative cells (Ki67high, white). Representative of 2 independent experiments. Scale bar: 20 μm. (j) qRT-PCR analysis of SMAD7, SNAI1 and STING expression in H2087-LCC cells treated with TGF-β or no additions for 2 h or 96 h, respectively. Mean ± s.e.m., representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (k) Representative IHC images (left) and quantification (right) of the proportion of H2087-LCC single cells, small clusters, micrometastases, and macrometastases in the lungs of NSG mice 7 weeks after intravenous injection of 1x105 cells. Scale bar: 50 μm. n = 102 (WT) or 33 (KO) single cells, 17 (WT) or 12 (KO) small clusters, 31 (WT) or 71 (KO) micrometastases, and 5 (WT) or 9 (KO) macrometastases. (l) IF staining of human vimentin (green), DAPI (blue), and CCL5 (red), in lung metastases generated 7 weeks after intravenous inoculation of 1x105 WT or TGFBR2 knockout H2087-LCC cells in NSG mice. The IF staining intensity in vimentin-positive areas was quantified. Scale bar: 50 μm. n = 41 (WT) or 55 (KO) lesions from 5 mice. Mean ± s.e.m., two-sided unpaired t-test.
Extended Data Fig. 11 Immune cell dependence of STING agonist-mediated suppression of metastasis reactivation.
(a) Rationale for experimental design. (b-c) qRT-PCR analysis of CXCL10 and CCL5 mRNA levels in WT or STING knockout KPad1 (b) or H2087-LCC (c) cells treated with 33 μM MSA-2 for 4 h. Mean ± s.e.m., representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (d–e) Treatment was performed as in (b, c) but with 10 μM (KPad1) or 50 μM (H2087-LCC) ADU-S100. Mean ± s.e.m., representative of 2 independent experiments. Each dot represents a technical replicate of the assay. (f-j) Metastasis-free survival plots of B6-albino mice intracardially inoculated with 2.5x105 KPad1 cells, treated with both MSA-2 and IgG control (f, n = 10 mice) or individual antibodies to deplete NK cells (g, n = 10 mice for vehicle or 8 mice for MSA-2), CD4+ T cells (h, n = 10 mice), CD8+ T cells (i, n = 10 mice), or a combination of these antibodies (j, n = 10 mice for vehicle or 9 mice for MSA-2). Mice were administered antibodies (200 μg/mouse) once weekly for 3 weeks starting 6 days after cell inoculation, and vehicle or MSA-2 (50 mg/kg of body weight) once weekly for 2 weeks starting 9 days after cell inoculation. Log-rank test. (k) Metastasis-free survival plots of B6-albino mice intracardially inoculated with 2.5 x 105 KPad1 cells pre-treated with 33 μM MSA-2 for 24h before inoculation. n = 10 mice per group. (l) Metastasis-free survival plots of C57BL/6J mice intravenously inoculated with 2.5 x 105 WT or Sting knockout KPad1 cells and treated with vehicle or ADU-S100 (1.25 mg/kg of body weight) once weekly by intratracheal delivery until the study endpoint. ADU-S100 treatment started 5 days after cell inoculation. n = 15 mice (control) or 8 mice (ADU-S100). Log-rank test. (m) Quantification of WT or STING knockout H2087-LCC cell numbers in lungs from athymic mice treated with vehicle or ADU-S100 (6.25 mg/kg of body weight) once weekly for 4 weeks by intratracheal delivery. ADU-S100 treatment started 1 week after intravenous inoculation of 1 x 105 cells. Lungs were harvested 5 weeks after inoculation. n = 5 mice per group. Mean ± s.e.m., two-sided unpaired t-test.
Supplementary information
Supplementary Figures
This file contains two supplementary figures. Supplementary Fig. 1. Raw images of uncropped gels. Supplementary Fig. 2. Example of gating strategy for flow cytometry analysis of cDC1 cells, NK cells and T cells.
Supplementary Table 1
GSEA analysis showing significantly enriched hallmarks in proliferative H2087-LCC cells compared to immune-evasive metastatic outbreaks, analysed from scRNA seq data (Laughney et al., 2020).
Supplementary Table 2
sgRNA sequences for CRISPR screens of human H2087-LCC.
Supplementary Table 3
sgRNA sequences for CRISPR screens of mouse KPad1.
Supplementary Table 4
Metastatic potential to the indicated organs from H2087-LCC and KPad1 cells expressing the individual sgRNA pools.
Supplementary Table 5
CRISPR screen score in H2087-LCC and KPad1.
Supplementary Table 6
Gene signatures.
Supplementary Table 7
CpG sites in STING promoter and 3′ enhancer.
Supplementary Table 8
Primers for CRISPR screen.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, J., Sánchez-Rivera, F.J., Wang, Z. et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature 616, 806–813 (2023). https://doi.org/10.1038/s41586-023-05880-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-05880-5
This article is cited by
-
Role of STING in the treatment of non-small cell lung cancer
Cell Communication and Signaling (2024)
-
cGAS-STING pathway expression correlates with genomic instability and immune cell infiltration in breast cancer
npj Breast Cancer (2024)
-
LncRNA Malat1 suppresses pyroptosis and T cell-mediated killing of incipient metastatic cells
Nature Cancer (2024)
-
Harnessing innate immune pathways for therapeutic advancement in cancer
Signal Transduction and Targeted Therapy (2024)
-
Second messenger 2'3'-cyclic GMP-AMP (2'3'-cGAMP): the cell autonomous and non-autonomous roles in cancer progression
Acta Pharmacologica Sinica (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.