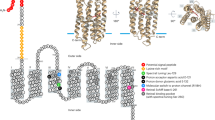
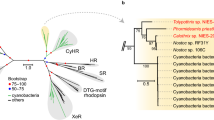
Abstract
Energy transfer from light-harvesting ketocarotenoids to the light-driven proton pump xanthorhodopsins has been previously demonstrated in two unique cases: an extreme halophilic bacterium1 and a terrestrial cyanobacterium2. Attempts to find carotenoids that bind and transfer energy to abundant rhodopsin proton pumps3 from marine photoheterotrophs have thus far failed4,5,6. Here we detected light energy transfer from the widespread hydroxylated carotenoids zeaxanthin and lutein to the retinal moiety of xanthorhodopsins and proteorhodopsins using functional metagenomics combined with chromophore extraction from the environment. The light-harvesting carotenoids transfer up to 42% of the harvested energy in the violet- or blue-light range to the green-light absorbing retinal chromophore. Our data suggest that these antennas may have a substantial effect on rhodopsin phototrophy in the world’s lakes, seas and oceans. However, the functional implications of our findings are yet to be discovered.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available in the main text or the Supplementary Information. The sequence of fosmid Kin4B8 was deposited in GenBank under accession number OP056329. Atomic coordinates of the crystal structure of Kin4B8 have been deposited in the Protein Data Bank under 7YTB. The density map and structure coordinate of the cryo-EM structure of the zeaxanthin-bound Kin4B8 have been deposited in the Electron Microscopy Data Bank and the Protein Data Bank with accession numbers EMD-35143 and 8I2Z, respectively. Source data are provided with this paper.
Code availability
The code used for the bioinformatic analyses is available from the GitHub repository (https://github.com/BejaLab/antenna) and the data are deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.20502384).
References
Balashov, S. P. et al. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309, 2061–2064 (2005).
Imasheva, E. S., Balashov, S. P., Choi, A. R., Jung, K.-H. & Lanyi, J. K. Reconstitution of Gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry 48, 10948–10955 (2009).
Fuhrman, J. A., Schwalbach, M. S. & Stingl, U. Proteorhodopsins: an array of physiological roles? Nat. Rev. Microbiol. 6, 488–494 (2008).
Vollmers, J. et al. Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS ONE 8, e63422 (2013).
Bertsova, Y. V., Arutyunyan, A. M. & Bogachev, A. V. Na+-translocating rhodopsin from Dokdonia sp. PRO95 does not contain carotenoid antenna. Biochem. Mosc. 81, 414–419 (2016).
Misra, R., Eliash, T., Sudo, Y. & Sheves, M. Retinal–salinixanthin interactions in a thermophilic rhodopsin. J. Phys. Chem. B 123, 10–20 (2019).
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000).
Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001).
Atamna-Ismaeel, N. et al. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2, 656–662 (2008).
Frigaard, N.-U., Martinez, A., Mincer, T. J. & DeLong, E. F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439, 847–850 (2006).
Finkel, O. M., Béjà, O. & Belkin, S. Global abundance of microbial rhodopsins. ISME J. 7, 448–451 (2013).
Gómez-Consarnau, L. et al. Microbial rhodopsins are major contributors to the solar energy captured in the sea. Sci. Adv. 5, eaaw8855 (2019).
DeLong, E. F. & Béjà, O. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 8, e1000359 (2010).
Munson-McGee, J. H. et al. Decoupling of respiration rates and abundance in marine prokaryoplankton. Nature 612, 764–770 (2022).
Wang, W.-W., Sineshchekov, O. A., Spudich, E. N. & Spudich, J. L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 278, 33985–33991 (2003).
Man, D. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725–1731 (2003).
Lanyi, J. K. & Balashov, S. P. in Halophiles and Hypersaline Environments (eds. Ventosa, A., Oren, A. & Ma, Y.) 319–340 (Springer, 2011).
Balashov, S. P. et al. Reconstitution of Gloeobacter rhodopsin with echinenone: role of the 4-keto group. Biochemistry 49, 9792–9799 (2010).
Kopejtka, K. et al. A bacterium from a mountain lake harvests light using both proton-pumping xanthorhodopsins and bacteriochlorophyll-based photosystems. Proc. Natl Acad. Sci. USA 119, e2211018119 (2022).
Pushkarev, A. & Béjà, O. Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J. 10, 2331–2335 (2016).
Pushkarev, A. et al. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595–599 (2018).
Chazan, A. et al. Diverse heliorhodopsins detected via functional metagenomics in freshwater Actinobacteria, Chloroflexi and Archaea. Environ. Microbiol. 24, 110–121 (2022).
Inoue, K. et al. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013).
Bhosale, P. & Bernstein, P. S. Microbial xanthophylls. Appl. Microbiol. Biotechnol. 68, 445–455 (2005).
Demmig-Adams, B., Polutchko, S. K. & Adams, W. W. Structure–function–environment relationship of the isomers zeaxanthin and lutein. Photochem 2, 308–325 (2022).
Barreiro C. & Barredo J. L. Microbial Carotenoids: Methods and Protocols (Humana Press, 2018).
Ram, S., Mitra, M., Shah, F., Tirkey, S. R. & Mishra, S. Bacteria as an alternate biofactory for carotenoid production: a review of its applications, opportunities and challenges. J. Funct. Foods 67, 103867 (2020).
Shibata, M. et al. Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 8, 8262 (2018).
Luecke, H. et al. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl Acad. Sci. USA 105, 16561–16565 (2008).
Chuon, K. et al. Assembly of natively synthesized dual chromophores into functional actinorhodopsin. Front. Microbiol. 12, 652328 (2021).
Yoshizawa, S., Kawanabe, A., Ito, H., Kandori, H. & Kogure, K. Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ. Microbiol. 14, 1240–1248 (2012).
Ahmed, F. et al. Profiling of carotenoids and antioxidant capacity of microalgae from subtropical coastal and brackish waters. Food Chem. 165, 300–306 (2014).
Shihoya, W. et al. Crystal structure of heliorhodopsin. Nature 574, 132–136 (2019).
Kishi, K. E. et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689.e23 (2022).
Balashov, S. P., Imasheva, E. S., Wang, J. M. & Lanyi, J. K. Excitation energy-transfer and the relative orientation of retinal and carotenoid in xanthorhodopsin. Biophys. J. 95, 2402–2414 (2008).
Lakowicz, J. R. (ed.) in Principles of Fluorescence Spectroscopy 27–61 (Springer, 2006).
Dana, J. et al. Testing the fate of nascent holes in CdSe nanocrystals with sub-10 fs pump–probe spectroscopy. Nanoscale 13, 1982–1987 (2021).
Polívka, T. et al. Femtosecond carotenoid to retinal energy transfer in xanthorhodopsin. Biophys. J. 96, 2268–2277 (2009).
Iyer, E. S. S., Gdor, I., Eliash, T., Sheves, M. & Ruhman, S. Efficient femtosecond energy transfer from carotenoid to retinal in Gloeobacter rhodopsin–salinixanthin complex. J. Phys. Chem. B 119, 2345–2349 (2015).
Doi, S., Tsukamoto, T., Yoshizawa, S. & Sudo, Y. An inhibitory role of Arg-84 in anion channelrhodopsin-2 expressed in Escherichia coli. Sci. Rep. 7, 41879 (2017).
Nagiri, C. et al. Crystal structure of human endothelin ETB receptor in complex with peptide inverse agonist IRL2500. Commun. Biol. 2, 236 (2019).
Yamashita, K., Hirata, K. & Yamamoto, M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. D Struct. Biol. 74, 441–449 (2018).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine.Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3.eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D Struct. Biol. 77, 1282–1291 (2021).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Inoue, K. et al. Exploration of natural red-shifted rhodopsins using a machine learning-based Bayesian experimental design. Commun. Biol. 4, 362 (2021).
Salazar, G. et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179, 1068–1083.e21 (2019).
Chen, I.-M. A. et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763 (2021).
Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021).
Sunagawa, S. et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 10, 1196–1199 (2013).
Wickham, H. in ggplot2 (eds Gentleman, R., Hornik, K. & Parmigiani, G.) 189–201 (Springer, 2016).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Acknowledgements
We thank J. K. Lanyi for commenting on the manuscript; M. Shalev-Benami for help in optimizing protein purification; Y. Ferguson for help in figure design; G. Tzuri and T. Isaacson for sharing materials; J. Anton for providing S. ruber for the isolation of salinixanthin standard; E. Martinez-Montes, director of the ‘Marismas del Odiel’ Natural Park, for support in obtaining the Atlantic Sea water sample; the Yigal Allon Kinneret Limnological Laboratory (KLL) for technical help with Lake Kinneret sampling; the Interuniversity Institute for Marine Sciences (IUI) in Eilat for providing access to their pier as well as workspace for the primary processing of the Red Sea samples; and the Israel Nature and Parks Authority for the permit to sample in Ein Afek reserve. This work was supported by the Israel Science Foundation (grant 3592/19 to O.B.), the Institute for Fermentation Osaka (W.S.), JSPS KAKENHI (grants 18H04136 and 22H00557 to S.Y., JP21H01875 and JP20K21383 to K.I., 19H05777 to W.S., 21H04969 to H.K. and 21H05037 to O.N.), MEXT Advancement of Technologies for Utilizing Big Data of Marine Life (grant JPMXD1521474594 to S.Y.), MEXT KAKENHI, Grant-in-Aid for Transformative Research Areas (B) ‘low-energy manipulation’ (grant JP20H05758 to K.I.), the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from the Japan Agency for Medical Research and Development (AMED) under grant number JP19am0101070 (support number 1627 to W.S.), Agencia Estatal de Investigación/FEDER, UE (grant number 2019-110438RB-C22 to R.L.), and Binational Science Foundation (grants 2016102 and 2020105 to S.R.). S.R. holds the Lester Aronberg Chair in Chemistry. M.S. holds the Katzir-Makineni Chair in Chemistry. O.B. holds the Louis and Lyra Richmond Chair in Life Sciences.
Author information
Authors and Affiliations
Contributions
A.C. and A.P. conceived the project, performed environmental sampling and functional metagenomics. A.C. performed carotene extraction, protein biochemistry, carotene binding and light-dependent proton pumping. A.R. performed bioinformatics. S.L. performed molecular biology. I.D. and M.S. performed absorption, emission and CD spectroscopies. T.F., M.H., Y.T. and S.Y. performed absorption, emission and carotene characterization of the PR-containing flavobacterial isolate. S.M., F.K.S., T.T., W.S. and O.N. performed structural analysis. A.M.-M., P.G.-V. and R.L. performed carotene characterization from environmental samples and from rhodopsin-bound carotenes. P.M. and S.R. performed ultrafast spectroscopy. T.I., M.K., T.N. and K.I. performed laser-flash photolysis. Y.M., K.K., R.A.-Y. and H.K. performed low-temperature UV–Vis and FTIR spectroscopy. O.B. coordinated the project. A.C. and O.B. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Valentin Gordeliy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of Lake Kinneret and Atlantic coast chromophore extracts.
a, HPLC profile of Lake Kinneret chromophore extract. Main peaks correspond to myxoxanthophyll (1), dinoxanthin (2), diatoxanthin (3), lutein (4), zeaxanthin (5), canthaxanthin (6), chlorophyll b (7), chlorophyll a (8), echinenone (9), pheophytin a (10), and β-carotene (11). b, HPLC profile of Atlantic coast chromophore extract. Main peaks correspond to chlorophyll c (1), siphonaxanthin (2), fucoxanthin (3), violaxanthin (4), diadinoxanthin (5), diatoxanthin (6), lutein (7), zeaxanthin (8), chlorophyll b (9), chlorophyll a (10), and β-carotene (11). Chromophores registered at 450 nm.
Extended Data Fig. 2 Spectroscopic characterization of diverse rhodopsins bound to lutein.
a and d, Absorbance change of different rhodopsins upon incubation with lutein. b, e, CD spectra with and without lutein. c, f, Fluorescence excitation spectra with and without lutein; emission monitored at 720 nm.
Extended Data Fig. 3 Quantum efficiency of excitation energy transfer (EET) from xanthophylls to different rhodopsins and ultrafast spectroscopy characterization of lutein in complex with Kin4B8.
a, Calculated quantum efficiency of EET from lutein and zeaxanthin to different rhodopsins in complex. b, Quantum efficiency of EET from lutein to different rhodopsins in complex, as a function of wavelength. c, S2 state decay of lutein. Blue — Kin4B8-lutein, black — Kin4B8 (reduced RPSB bond)-lutein, and red- their fit. X-axis presents the delay between pump and probe. Y-axis shows probe absorption difference in the presence and absence of pump pulse. Kinetic data were fitted with a function convolving 40 fs gaussian IRF and a mono-exponential decay. d, Reduction of retinal protonated Schiff-base by NaBH4 in Kin4B8-lutein complex. Blue — absorption spectrum of Kin4B8-lutein before reduction. Black — absorption spectrum of Kin4B8-lutein after reduction.
Extended Data Fig. 4 FTIR Influence of lutein on the retinal photoisomerization in Kin4B8 at 77 K.
a, UV-visible absorption spectra of lipid-reconstituted Kin4B8 with (top) and without (bottom) lutein at 77 K. One division of the y-axis corresponds to 0.5 absorbance units. b, Difference UV-visible spectra upon illumination of Kin4B8 with (top) and without (bottom) lutein. Hydrated films of lipid-reconstituted Kin4B8 were first illuminated at 540 nm light (solid lines), followed by illumination at >590 nm light (broken lines) at 77 K. Solid and broken lines are mirror-imaged, indicating photochromic properties for Kin4B8 and the K intermediate. One division of the y-axis corresponds to 0.05 absorbance units. c, Difference FTIR spectra upon illumination of Kin4B8 with (top) and without (bottom) lutein. Hydrated films of lipid-reconstituted Kin4B8 with H2O were first illuminated at 540 nm light (solid lines), followed by illumination at >590 nm light (dotted lines) at 77 K. One division of the y-axis corresponds to 0.002 absorbance units. d, e, Light-induced difference UV-visible (d) and FTIR (e) spectra of Kin4B8 with (red) and without (black) lutein, where positive and negative signals originate from the K intermediate and unphotolyzed Kin4B8, respectively.
Extended Data Fig. 5 Spectroscopic characterization of Kin4B8 and Kin4B8-G153F bound to carotenoids.
a, Absorption spectra of Kin4B8 with salinixanthin (Sal). b, CD spectra of Kin4B8 with and without salinixanthin. c, Fluorescence excitation spectra of Kin4B8 with and without salinixanthin; emission monitored at 720 nm. d, Absorption spectra of Kin4B8 with β-carotene (β-car). e, CD spectra of Kin4B8 with and without β-carotene.f, .Absorption spectra of Kin4B8-G153F with zeaxanthin (Zeax). e, CD spectra of Kin4B8-G153F with and without zeaxanthin.
Extended Data Fig. 6 The photocycle of Kin4B8.
a, Chromatogram of HPLC analyses (left) and the compositions of the retinal isomers (right) in Kin4B8 without (top) and with (bottom) lutein under the dark (gray) and light-adapted (green) conditions. at, 11, 13, syn, and anti indicate all-trans, 11-cis, 13-cis, syn, and anti configurations, respectively. b, Two-dimensional plot of transient absorption change (left), transient absorption spectra at different time points (middle), and time course of the transient absorption change (right) of Kin4B8 without (top) and with (bottom) lutein. Peaks derived from the absorption change of lutein are indicated by an asterisk. c, Absorption spectra of the photointermediates of Kin4B8 without (left) and with (right) lutein. Peaks derived from the absorption change of lutein are indicated by asterisks. d, Photocycle model of Kin4B8. Conformational change of rhodopsin affects the structure of lutein (blue arrow) from K3/M1 to O4/Kin4B8′2. e, The ratios of transient absorption change in Kin4B8 with and without lutein at different excitation wavelengths (415, 432, 457, 473, 487, 552, and 601 nm) (bars colored according to the color of excitation light). The absorption spectra of Kin4B8 without (pink line) and with (orange line) lutein were overlaid. The red dashed line indicates no difference between without and with lutein.
Extended Data Fig. 7 Light-induced proton-pumping activity of Kin4B8 and Kin4B8-G153F.
a and b, Monitoring of pH changes in E. coli spheroplasts suspension expressing Kin4B8 or Kin4B8-G153F, respectively, with and without zeaxanthin. The spheroplasts were illuminated with violet (430 nm), blue (450 nm), or green (550 nm) light for 2 min (indicated by the colored bars). An enlarged plot of the first 15 s of illumination is displayed to the right of each measurement. The presented traces are the average of six or more independent biological replicates (Error bars represent the SEM).
Extended Data Fig. 8 Diversity and distribution of PRs and XRs with (G) and without (FW) fenestration among different prokaryotic phyla across four environments.
a, Maximum likelihood phylogenetic analysis of the PR-XR-NQ clade based on representative protein sequences. Characterized ion pumps are indicated with dots, terminal branches are colored by the corresponding phylum. Major clades with more than one representative are highlighted and labeled. The tree is outgroup rooted. b, Distribution of PRs and XRs with the canonical TM3 motif DTE among genomes assigned to different taxa, with (G) and without (FW) fenestration. The analysis is based on GEM genomes and the numbers are summarized per operational taxonomic unit (OTU). The colors are as in panel (a). c, Relative abundance of different families of the clade across four habitats based on the metagenomic data from IMG/M. Only families with a total relative abundance of >0.1% are shown. d, Predicted absorption maxima for PRs and XRs with the three most frequent residues at the fenestration position. Individual observation corresponds to an average absorption maximum predicted with the rhodopsin BLASSO model for sequences with the same 24 residues of the retinal binding pocket56. The sequences from OM-RGC, IMG/M and GEM were pooled together. The size of the dots is proportional to the number of distinct rhodopsin domain sequences and the color approximates the predicted mean absorption spectra. Statistical differences between the groups were assessed with Dunn’s test with FDR correction. Significance levels are indicated with asterisks: *** – adjusted p-values < 0.001. Abbreviation of family names in (A) and (C): ACB – Archaea clade B, ESR – Exiguobacterium sibiricum rhodopsin, NQ – NQ sodium and chloride pumps, MACR – marine actinobacteria clade rhodopsins, PR – proteorhodopsins, TAT – TAT rhodopsins, XR – xanthorhodopsins, P1 – unnamed clade including QsActR, KrActR and related rhodopsins, P3 and P4 – currently unnamed clades.
Extended Data Fig. 9 Structural features of Kin4B8.
a, Cryo-EM single-particle analysis of the zeaxanthin-bound Kin4B8. b, Comparison of the cryo-EM and crystal structures of Kin4B8. c, Cryo-EM density of the zeaxanthin, which allows unambiguous identification of the molecule. In particular, resolution of the half of zeaxanthin proximal to the fenestration is high enough to permit identification of the dimethyl group of its hydroxyl ring. d, Comparison of the oligomeric structures of Kin4B8, S. ruber XR (PDB ID: 3DDL), BR (PDB ID: 1C3W), GPR (PDB ID: 7B03). With the ECL1 sheet inside, Kin4B8 forms a hexamer with aligned directions to the membrane in the crystal packing. The pentameric structure would reflect a physiological condition, in contrast to the previously reported head-to-tail dimer of S. ruber XR. e, Structural comparison of Kin4B8 with S. ruber XR (PDB ID: 3DDL), bacteriorhodopsin (BR) (PDB ID: 1C3W), and GPR (PDB ID: 7B03), with root mean square deviations (RMSD) of 1.44, 1.96, and 2.61 Å, respectively. Notably, the N-terminal region (residues 6–11) and ECL1 form a 3-stranded antiparallel β-sheet, as in S. ruber XR and other omega rhodopsins. f, Key rhodopsin proton pump motifs in Kin4B8. Black dashed lines indicate hydrogen-bonding interactions. Red spheres indicate water molecules.
Extended Data Fig. 10 Global distribution of fenestrated XRs and PRs based on the Tara Oceans data.
Rhodopsin sequences from the Ocean Microbial Reference Catalog v.2 were classified into PRs and XRs with the canonical DTE motif, and binned into those having bulky residues (Phe or Trp) or Gly at XR position 156. Individual pie charts represent ratios between fenestrated (yellow) and non-fenestrated (blue) PRs and XRs by location and the size of the charts is proportional to the total abundance of DTE PR- and XR-encoding ORFs (a) and this abundance relative to the abundance of ten single-copy markers (b).
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–2 and Supplementary Tables 1–2.
Supplementary Data
Source Data for Supplementary Fig. 2.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chazan, A., Das, I., Fujiwara, T. et al. Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Nature 615, 535–540 (2023). https://doi.org/10.1038/s41586-023-05774-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-05774-6
This article is cited by
-
Engineering artificial photosynthesis based on rhodopsin for CO2 fixation
Nature Communications (2023)
-
Plastid-localized xanthorhodopsin increases diatom biomass and ecosystem productivity in iron-limited surface oceans
Nature Microbiology (2023)
-
Responding to light signals: a comprehensive update on photomorphogenesis in cyanobacteria
Physiology and Molecular Biology of Plants (2023)
-
Expression of Xanthorhodopsin in Escherichia coli
The Protein Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.