Abstract
Oncoproteins of the MYC family drive the development of numerous human tumours1. In unperturbed cells, MYC proteins bind to nearly all active promoters and control transcription by RNA polymerase II2,3. MYC proteins can also coordinate transcription with DNA replication4,5 and promote the repair of transcription-associated DNA damage6, but how they exert these mechanistically diverse functions is unknown. Here we show that MYC dissociates from many of its binding sites in active promoters and forms multimeric, often sphere-like structures in response to perturbation of transcription elongation, mRNA splicing or inhibition of the proteasome. Multimerization is accompanied by a global change in the MYC interactome towards proteins involved in transcription termination and RNA processing. MYC multimers accumulate on chromatin immediately adjacent to stalled replication forks and surround FANCD2, ATR and BRCA1 proteins, which are located at stalled forks7,8. MYC multimerization is triggered in a HUWE16 and ubiquitylation-dependent manner. At active promoters, MYC multimers block antisense transcription and stabilize FANCD2 association with chromatin. This limits DNA double strand break formation during S-phase, suggesting that the multimerization of MYC enables tumour cells to proliferate under stressful conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing datasets generated during this study are available at the Gene Expression Omnibus under accessions GSE205585 and GSE183094. The 4sU-seq dataset of MYC-depleted cells is available under accession GSE115365. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE70 partner repository with the dataset identifier PXD036674.
Code availability
The code used for super-resolution microscopy analysis is available on GitHub at https://github.com/super-resolution/Solvie-et-al-supplement.
References
Dhanasekaran, R. et al. The MYC oncogene—the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19, 23–36 (2022).
Lourenco, C. et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 21, 579–591 (2021).
Baluapuri, A., Wolf, E. & Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21, 255–267 (2020).
Herold, S. et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567, 545–549 (2019).
Roeschert, I. et al. Combined inhibition of Aurora-A and ATR kinase results in regression of MYCN-amplified neuroblastoma. Nat. Cancer 2, 312–326 (2021).
Endres, T. et al. Ubiquitylation of MYC couples transcription elongation with double-strand break repair at active promoters. Mol. Cell. 81, 830–844.e813 (2021).
Saldivar, J. C., Cortez, D. & Cimprich, K. A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017).
Tarsounas, M. & Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell. Biol. 21, 284–299 (2020).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018).
Lorenzin, F. et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
Lin, C. Y. et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012).
Rosales, T. et al. Partition of Myc into immobile vs. mobile complexes within nuclei. Sci. Rep. 3, 1953 (2013).
Trader, D. J., Simanski, S. & Kodadek, T. A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J. Am. Chem. Soc. 137, 6312–6319 (2015).
Lamberto, I. et al. Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem. Biol. 24, 1490–1500.e1411 (2017).
Gregory, M. A., Qi, Y. & Hann, S. R. Phosphorylation by glycogen synthase kinase-3 controls c-Myc proteolysis and subnuclear localization. J. Biol. Chem. 278, 51606–51612 (2003).
Kaida, D. et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 (2007).
Carter, D. R. et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci. Transl. Med. 7, 312ra176 (2015).
Cugusi, S. et al. Heat shock induces premature transcript termination and reconfigures the human transcriptome. Mol. Cell 82, 1573–1588.e10 (2022).
Wang, H. et al. Improved low molecular weight Myc–Max inhibitors. Mol. Cancer Ther. 6, 2399–2408 (2007).
Li, Y., Ma, X., Wu, W., Chen, Z. & Meng, G. PML nuclear body biogenesis, carcinogenesis, and targeted therapy. Trends Cancer 6, 889–906 (2020).
Arabi, A., Rustum, C., Hallberg, E. & Wright, A. P. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J. Cell Sci. 116, 1707–1717 (2003).
Drygin, D. et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71, 1418–1430 (2011).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Lam, S. S. et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015).
van Riggelen, J. et al. The interaction between Myc and Miz1 is required to antagonize TGFβ-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 24, 1281–1294 (2010).
Baluapuri, A. et al. MYC recruits SPT5 to RNA polymerase ii to promote processive transcription elongation. Mol. Cell 74, 674–687.e611 (2019).
Papadopoulos, D. et al. MYCN recruits the nuclear exosome complex to RNA polymerase II to prevent transcription-replication conflicts. Mol. Cell 82, 159–176.e112 (2022).
Das, S. K. et al. MYC assembles and stimulates topoisomerases 1 and 2 in a “topoisome”. Mol. Cell 82, 140–158.e112 (2022).
Nikiforov, M. A. et al. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 22, 5054–5063 (2002).
Fuchs, M. et al. The p400 complex is an essential E1A transformation target. Cell 106, 297–307 (2001).
Hingorani, S. R. et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005).
Krenz, B. et al. MYC- and MIZ1-dependent vesicular transport of double-strand RNA controls immune evasion in pancreatic ductal adenocarcinoma. Cancer Res. 81, 4242–4256 (2021).
Chaudhury, I., Stroik, D. R. & Sobeck, A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 34, 3939–3954 (2014).
Dupre, A. et al. A forward chemical genetic screen reveals an inhibitor of the Mre11–Rad50–Nbs1 complex. Nat. Chem. Biol. 4, 119–125 (2008).
Qiao, X. et al. UBR5 is coamplified with MYC in breast tumors and encodes an ubiquitin ligase that limits MYC-dependent apoptosis. Cancer Res. 80, 1414–1427 (2020).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl Acad. Sci. USA 101, 9085–9090 (2004).
Diefenbacher, M. E. et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J. Clin. Invest. 124, 3407–3418 (2014).
Sun, X. X. et al. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc. Natl Acad. Sci. USA 112, 3734–3739 (2015).
Michel, M. A., Swatek, K. N., Hospenthal, M. K. & Komander, D. Ubiquitin linkage-specific affimers reveal insights into K6-linked ubiquitin signaling. Mol. Cell 68, 233–246.e235 (2017).
Heidelberger, J. B. et al. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 19, e44754 (2018).
He, X. et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 13, 1164–1171 (2017).
Hamperl, S. & Cimprich, K. A. Conflict resolution in the genome: how transcription and replication make it work. Cell 167, 1455–1467 (2016).
Yan, W. X. et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017).
Thomas, L. R. et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 58, 440–452 (2015).
Choe, K. N. et al. HUWE1 interacts with PCNA to alleviate replication stress. EMBO Rep. 17, 874–886 (2016).
Zhang, D., Zaugg, K., Mak, T. W. & Elledge, S. J. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126, 529–542 (2006).
de Vivo, A. et al. The OTUD5–UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 47, 729–746 (2019).
Kress, T. R., Sabo, A. & Amati, B. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 15, 593–607 (2015).
Gaillard, H., Garcia-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Meers, M. P., Bryson, T. D., Henikoff, J. G. & Henikoff, S. Improved CUT&RUN chromatin profiling tools. eLife 8, e46314 (2019).
Koulouras, G. et al. EasyFRAP-web: a web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 46, W467–W472 (2018).
Wolter, S. et al. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 (2012).
Kock, B. et al. Confinement and deformation of single cells and their nuclei inside size-adapted microtubes. Adv. Healthcare Materials 3, 1753–1758 (2014).
Kiskowski, M. A., Hancock, J. F. & Kenworthy, A. K. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 97, 1095–1103 (2009).
Cho, K. F. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999 (2020).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Li, J. et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 17, 399–404 (2020).
Zhang, L. & Elias, J. E. Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol. Biol. 1550, 185–198 (2017).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Quinlan, A. R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics https://doi.org/10.1002/0471250953.bi1112s47 (2014).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Petelski, A. A. et al. Multiplexed single-cell proteomics using SCoPE2. Nat. Protoc. 16, 5398–5425 (2021).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
This work was supported by Grants from the European Research Council (AuroMYC to M.E., TarMYC to E.W. and ULTRARESOLUTION to M.S.), the German Cancer Aid (no. 70113870 to M.E.), the German Research Foundation (WO 2108/1-1 to E.W. EI 222/21-1 to M.E., BA 7941/1-1 to A.B., GRK 2243 Ubiquitin to M.E. and E. W., and GRK 2157 to M.S. and M.J.). We thank C. Ade for sequencing, G. Cossa for performing the small compound screen, Y. Cruz García for the U2OS-MYC-AID cell line and T. Roth and A. Kutschke for technical support.
Author information
Authors and Affiliations
Contributions
D.S., A.B., L.U., D.F., T.E., A.A. and A.G. performed most of the experiments. D.P. provided unpublished data. I.M. and E.I. performed proteomic analyses. C.G. and J.J. performed DNA fibre assays. C.S.-V. performed high-content immunofluorescence experiments. M.J. performed dSTORM. T.K. performed structured illumination microscopy and FRAP experiments. S.D. performed dSTORM data analysis. A.B. analysed sequencing data. H.M. analysed antibody specificity. M.S., P.B., A.R., M.D., E.W. and M.E. devised and supervised experiments and E.W. and M.E. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
E.W. and M.E. are founders and stockholders of Tucana Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks George-Lucian Moldovan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Controls for dSTORM analyses.
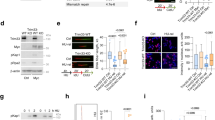
a. Heatmap of tiled MYC peptide microarrays. Arrays spotted with 30 amino acid MYC-peptides covering the complete protein were incubated with the indicated antibodies. The inset shows the sequenced recognized by the rabbit monoclonal Y69 antibody. b. Heatmap of tiled peptide microarrays for MYC, MYCN and MYCL. Arrays spotted with 15mer MYC-peptides covering the complete proteins were incubated with the Y69 antibody. c. dSTORM using MYC antibody Y69 in unperturbed U2OS cells at highly dilute labeling conditions. The inset shows a zoom-in in which individual signals can be distinguished. The staining was performed 4 times and each time 3–5 images were taken. Scale bar: 5 µm. d. Cumulative density functions of MYC molecules per cluster in unperturbed U2OS cells under saturating (dark blue) and diluted antibody (orange) labeling conditions and in data simulating a single-molecule distribution for saturating antibody concentrations (green) (n = 7 cells examined over 3 independent experiments). e. Percentage of MYC molecules in clusters of ≥ 100 molecules per cluster in unperturbed HCT116, HeLa, MCF10A and U2OS cells (n = 7 cells per cell line examined over 3 independent experiments). f. dSTORM using MYC antibody Y69 in U2OSMYC-Tet-On cells (DOX, 1 µg/ml, 24 h) at saturated labeling conditions. Staining was performed thrice with at least 2–4 images taken each time. Scale bar: 5 µm. g. Average Ripley`s K function for experimental (blue) and simulated (grey) localization data in unperturbed U2OS cells under diluted (left) and saturating (middle) labeling conditions, as well as in U2OSMYC-Tet-On cells (DOX, 1 µg/ml, 24 h) under saturating labeling conditions (right). In each panel the upper simulation represents the apparent clustering data expected for a monomeric protein distribution. The lower curve represents a homogeneous signal distribution of all localizations. 95% confidence intervals are colored in light grey. In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 2 Characterization of MYC Multimers.
a. Bar plot showing the results of a small molecule screen in U2OSMYC-Tet-On cells (DOX, 1 µg/ml, 24 h) to identify conditions that influence the formation of MYC multimers. Treatment with indicated compounds for 4 h. Bars below indicate grouping according to targets (DDR: DNA damage response, TOPO: Topisome). At minimum, 500 cells were analyzed in each condition per replicate. Shown are Z-scores for biological replicates with mean ± S.E.M. (n = 4). Mann-Whitney U, two-sided, with following p-values: MG-132 = 5e-5, RA-190 = 9.3e-4, P22077 = 3.9e-3, CBL0137 = 3.9e-3, PlaB = 9.3e-4. Inset shows representative example of automated spot detection. b. Validation using siRNAs in U2OSMYC-Tet-On cells (DOX, 1 µg/ml, 24 h). Multimers per nucleus assessed by immunofluorescence. Shown are Z-score normalized numbers of multimers. n = 1500 cells per condition examined over three independent experiments. Data are expressed as mean ± S.E.M. Two sided Mann-Whitney U test was used for comparison to non-targeting control (NTC) with following p-values: SF3B1 < 2.2e-16, PSMA3 < 2.2e-16, SUPT16H < 2.2e-16 (left). Exemplary pictures (right). Scale bar: 10 µm. c. Heatmap assessing influence of different stressors on MYC multimerization (FCCP: 5 µM, 18 h; puromycin 2.5 µg/ml,4 h; sucrose 0.2 M, 2 h; arsenite 0.5 mM,1 h; Q or Glu starvation 18 h; heat shock (HS) 30 min 42 °C). Multimers per nucleus assessed by immunofluorescence and normalized using Z-score. Mean of triplicates shown with ≥ 200 cells per condition per replicate (n = 3). d. Heatmap assessing MYC levels across a panel of indicated cell lines (transformed and non-transformed) treated as indicated (MG-132: 20 µM, 4 h) (left). Heatmap assessing MYC multimerization treated with MG-132 (20 µM), CBL0137 (5 µM) or PlaB (1 µM) for 4 h and HS (42 °C, 2 h). Where indicated, DOX was added to U2OSMYC-Tet-On cells for 24 h. Shown are mean of biological triplicates. Z-scores calculated within cell lines (n = 3) (right). e. Exemplary pictures of transformed and non-transformed cells upon indicated treatments. KPC: a KRAS/P53mut-driven pancreatic tumor cells32. Scale bar: 10 µm. f. Immunofluorescence of MYCN in IMR5 MYCN-amplified neuroblastoma cells upon 4 h treatment with MG-132 (20 µM), CBL0137 (5 µM) and PlaB (1 µM). Scale bar: 10 µm (n = 3). g. Confocal immunofluorescence images showing reversal of foci formation by MYC with 1,6-hexanediol following treatment with DOX (1 µg/ml, 24 h) and MG-132 (20 µM, 4 h) in U2OSMYC-Tet-On cells. Scale bar: 10 µm (n = 3). h. Immunofluorescence of U2OSMYC-Tet-On cells treated with DOX (1 µg/ml, 24 h), MG-132 (20 µM), CBL0137 (5 µM) and PlaB (1 µM) for 4 h or HS (42 °C for 30 min). Data are mean ± S.E.M. normalized to t = 0 (n = 3). Lines represent a polynomial regression model of 2nd-order. In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 3 Further characterization of MYC Multimers.
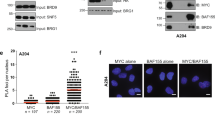
a. Immunoblots of co-IP experiments with tagged MYC proteins. MYC-FLAG and MYC-HA were transfected into HEK293T cells. MYC-FLAG was precipitated, and co-precipitated MYC-HA was analyzed by immunoblotting. Beads coupled to non-specific IgG, or cells expressing only MYC-HA were used as controls. VINCULIN shown as loading control (n = 3). For gel source data, see Supplementary Fig 1. b. PLAs to probe for multimeric MYC in sections from iliac crest of patients with myeloma/plasmacytoma. (Middle and right): The patient was treated with proteasomal inhibitor bortezomib in combination with radiotherapy, and normal hematopoietic cells are compared with myeloma cells. (Left): “CTL” are pretreatment biopsies. Scale bar: 10 µm (n = 2). c. Fluorescence pictures of in vitro droplet formation by recombinant MYC-mCherry (top) and ∆N-MYC-mCherry (bottom) (2, 1, 0.5, 0.25 µM protein) under different salt conditions (500, 250, 125 mM NaCl). Scale bar: 50 µm (n = 3). d. Representative images of in vitro assays probing for the permeability of MYC droplets for dextrans of different molecular weights. e. Pixelwise Pearson correlation coefficient of permeability measured by the mean fluorescence intensity of FITC-dextran detected within MYC droplets. Shown is mean ± S.E.M. (n = 3). f. Graph of predicted disordered domain (PONDR, VSL-2) of the MYC protein with indications of depleted or substituted regions in the MYC-mutants used. g. Bar graph visualizing multimerization ability of MYC-mutants compared to wildtype MYC(WT) upon MG-132 (20 µM, 4 h) treatment. Shown is the mean ± S.D. (n = 3). Unpaired t-test, two-sided, was used for comparison to full-length MYC (FL) with following p-values: ∆N-MYC = 6.1e-4, K-less-MYC = 6.39e-3, ∆TAD-MYC = 1.71e-3, ∆Box-MYC = 9.66e-2, ∆BoxII-MYC = 5.37e-1, ∆C-MYC = 3.47e-2, ∆BoxI-MYC = 2.15e-2. h. Fluorescence pictures of in vitro colocalization of recombinant MYC-mCherry (1 µM) with MAX-eGFP (1 µM) proteins. Controls only have single component. Scale bar: 50 µm (n = 3). In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 4 Relationship of MYC multimers to nucleoli and PML bodies.
a. Immunofluorescence of MAX and MYC upon treatment with DOX (1 µg/ml, 24 h) and MG-132 (20 µM, 4 h) in U2OSMYC-Tet-On cells. Scale bar: 5 µm (n = 3). b. Immunofluorescence of MAX and MYC upon treatment with DOX (1 µg/ml, 24 h), MG-132 (20 µM, 4 h) and 10058-F4 (150 µM, 6 h) in U2OSMYC-Tet-On cells. Scale bar: 5 µm (n = 3). c. Bar plot quantifying results from (b). Shown are fold changes of numbers of multimers compared to MG-132 treated cells. The number of cells analyzed are: DMSO (n = 494), MG-132 (n = 825), 10058-F4 (n = 1087), MG-132+10058-F4 (n = 852) over three independent experiments. Data are shown as mean ± S.E.M. Two sided Mann-Whitney U test was used for comparison to MG-132 treated cells with following p-values: DMSO < 2.2e-16, MG-132+10058-F4 < 2.2e-16. d. dSTORM of MYC using antibody Y69 in U2OS cells treated MG-132 (20 µM), CBL0137 (5 µM) and PlaB (1 µM) for 4 h or HS (42 °C for 30 min), at saturated labeling conditions (n = 3). Scale bar: 5 µm. e. Confocal immunofluorescence using RNA Polymerase I (RNAPI), PML (PCC = 0.465) and MYC in U2OSMYC-Tet-On cells treated with DOX (1 µg/ml, 24 h) and MG-132 (20 µM, 4 h). Scale bar: 5 µm (n = 3). f. Immunofluorescence of MYC and Fibrillarin upon treatment with DOX (1 µg/ml, 24 h), MG-132 (20 µM, 4 h) or CX5461 (0.5 µM, 4 h) in U2OSMYC-Tet-On cells. Scale bar: 5 µm (n = 3). In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 5 Redistribution of MYC on chromatin.
a. Heatmap of MYC occupancy analyzed by ChIP-Rx at 9,743 MYC promoters and 14,855 non-promoter sites in U2OSMYC-Tet-On treated as indicated (HS: 42 °C for 30 min; DOX: 1 µg/ml, 24 h) followed by recovery for 30 min and 5 h (n = 2). b. Proportion of total called MACS2 peaks of indicated proteins which show reduced (loss) or increased (gain) occupancy after 4 h MG-132 treated for MYC in U2OSMYC-Tet-On cells in absence or presence of DOX (CTL: n = 2; DOX: n = 3). c. Accumulation of JUN at 4,452 JUN-bound promoters documented by CUT&RUN. Average density plot of U2OSMYC-Tet-On cells at indicated conditions (MG-132: 20 µM, 4 h; DOX: 1 µg/ml, 24 h) (n = 1). d. MYC ChIP qPCR of chromatin from U2OSMYC-Tet-On cells documenting occupancy of MYC at the indicated promoters in control cells and exposure to DOX (1 µg/ml, 24 h), MG-132 (20 µM, 4 h), CBL0137 (5 µM, 5 h), or PlaB (1 µM, 4 h). Shown is mean of technical triplicates of one representative experiment of three biological replicates. e. Consensus motifs in non-promoter MYC binding sites using de-novo search algorithm (HOMER) at the 4,210 MYC binding sites non-promoters with increasing MYC levels and 6,644 promoter sites where MYC occupancy is reduced upon MG-132 (20 µM, 4 h) treatment (n = 2). f. Average density plot of MAX occupancy analyzed by CUT&RUN. MAX occupancy in U2OSMYC-Tet-On cells upon treatment with DOX (1 µg/ml, 24 h) and MG-132 (20 µM, 4 h) at 6,644 promoter sites where MYC occupancy is reduced (left) and 4,210 non-promoter sites with increasing MYC occupancy (right) upon MG-132 treatment (n = 1). g. Venn diagram of results from co-localization studies using quantitative immunofluorescence considering proteins as colocalizing at a cut-off of PCC = 0.3. h. GO-term analysis of significantly (FDR < 0.05) enriched proteins upon MYC multimerization in APEX2 experiments (Fig. 2c). Hypergeometric p-values with correction for multiple testing using the gSCS algorithm in gprofiler2. i. Average density plot of SPT5 ChIP-Rx across genes at the 6,644 promoter sites where MYC occupancy is reduced upon MG-132 treatment in U2OSMYC-Tet-On cells (MG-132: 20 µM, 4 h; DOX: 1 µg/ml, 24 h) (n = 2). In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 6 Functions of MYC multimers.
a. Volcano Plot of 4sU data showing transcriptional responses to changes in MYC levels (DOX: 1 µg/ml, 10 h) in U2OSMYC-Tet-On cells in presence or absence of MG-132 (20 µM, 5 h). Orange color indicates regulation +/− 1.5-fold. The color scale represents q-values for Benjamini-Höchberg multiple hypothesis correction (n = 3). b. Heatmap of Z-scores calculated from normalized 4sU Seq reads over 13,101 expressed genes, sorted for MYC driven gene regulation indicating transcriptional responses to doxycycline-mediated induction of MYC in the absence (top) and presence (bottom) of MG-132 (10 µM; 4 h) (n = 3). c. Pixel-wise Pearson correlation coefficient of pS2RNAPII and SPT5 calculated based on immunofluorescence data across the nucleus in absence (CTL) or presence of multimers (PlaB, 4 h, 1 µM; CBL0137, 4 h, 5 µM). n = 15 cells were analyzed per condition over three independent experiments. Box plots are defined as in Fig. 3g. Two sided Mann-Whitney U test was used. d. Illustration of the positional co-localization workflow (top). Positional co-localization of indicated proteins with MYC multimers in U2OSMYC-Tet-On cells upon treatment with DOX (1 µg/ml, 24 h) and MG-132 (20 µM, 4 h). Inner circle indicating the proportion of MYC multimers co-localizing with indicated protein. Outer circle indicating proportion of MYC multimers with homogeneous shape or hollow sphere (bottom). e. Confocal Immunofluorescence pictures of unperturbed U2OSMYC-Tet-On cells of indicated proteins. Scale bar: 5 µm (n = 3). f. Immunofluorescence pictures of U2OSMYC-Tet-On cells showing MYC and pATR upon treatment with DOX (1 µg/ml, 24 h), MG-132 (20 µM, 4 h) and CX-5461 (1 µM, 4 h). Scale bar: 5 µm (n = 3). g. Confocal imaging of triple staining of MYC, BRCA1 and Fibrillarin upon MG-132 treatment (4 h, 20 µM) Scale bar: 5 µm (n = 3). In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 7 Characterization of MYC binding sites on chromatin.
a. Heatmap from CUT&RUN sequencing data showing FANCD2 (n = 2) and ATR (n = 1) association with (left) non-promoter and (right) promoter sites in control U2OS cells and in cells exposed to hydroxyurea (HU, 5 mM, 2 h) or Aphidicolin (APH, 400 nM, 14 h) as indicated. Heatmap centered around peaks called in FANCD2 treated with APH. b. Correlation of change in MYC occupancy after MG-132 or HU treatment at n = 31,970 peaks called from FANCD2 CUT&RUN in one of two independent experiments with similar results (HU: 3.5 mM, 4 h; MG-132: 20 µM, 4 h) (p-value < 2.2*10−16; significance level of Pearson’s product-moment correlation t-test, two-sided). c. Heatmaps showing input-normalized MYC ChIP-Rx reads, RNAPII ChIP-Rx sequencing reads or FANCD2 and ATR CUT&RUN reads in U2OS cells treated with MG-132 (20 µM, 5 h), Aphidicolin (APH, 0.4 µM, 14 h) or hydroxyurea (HU, 5 mM, 2 h) of 909 promoters with MYC peaks which have FANCD2 and ATR peaks upstream. d. Normalized 4sU reads at MYC non-promoter sites (4,270) in MYC-depleted (left, siMYC; 48 h) and in MG-132 treated cells (right, 20 µM, 4 h) relative to control cells (n = 3). e. Heatmap of reads from ATR CUT&RUN indicating chromatin occupancy in U2OSMYC-Tet-On cells. Identical positions as used in Fig. 3i (1,478 promoter sites) (n = 2). f. Average density plots of FANCD2 (left) & MYC (right) CUT&RUN in PDAC cells in presence (shMYC) or absence of an shRNA targeting MYC (CTL) at 14,295 FANCD2 sites upon HU treatment, 3.5 mM for 4 h) (n = 1). g. Scheme of fiber assay experiments with indicated experimental time points to assess for degradation (T2) and restart (T3). h. Fiber assay assessing fork degradation (T2) upon MYC depletion by shRNA (1 µg/ml DOX, 24 h) in KPC cells treated as illustrated in (g). Only fibers containing three labels (red-green-red) were considered. Mean second CldU track length (µm) of one of three biological replicates with similar results is shown ± S.E.M. P-values (Mann-Whitney U test, two-sided) are for n = 142 (CTL), n = 102 (DOX), n = 141 (CTL+Mirin) and n = 109 (DOX+Mirin) fibers. i. Fiber assay assessing fork restart (T3). Percentage of fibers that incorporation of the fourth label was quantified over the total number of fibers, which contained three (red-green-red). Data show mean ± S.D. (n = 3) with each replicate quantifying more than 500 fibers per condition (CTL: 534, 515, 527; DOX: 537, 557, 606). p-value: unpaired t-test, two-sided. In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 8 Control of MYC multimerization by ubiquitylation and SUMOylation.
a. Confocal immunofluorescence of indicated proteins in U2OS cells expressing WT MYC or K-less MYC. Treated as in Fig. 2d. Scale bar: 20 µm (n = 3). b. MYC-MYC PLA in U2OSMYC-AID cells reconstituted with WT MYC or K-less MYC as indicated. Endogenous MYC depleted by addition of indoleacetic acid (500 µM, 6 h). PLA (red) is overlaid with nuclear staining (blue) and compared with MYC staining (green). Scale bar: 5 µm (n = 3). c. Immunoblot of endogenous K6-linked ubiquitin immunoprecipitations from U2OS cell extracts. Immunoprecipitated K6-linked ubiquitin was visualized using a α-GFP antibody detecting the K6 affimer. Actin was used as a loading control, *IgG heavy chain (n = 1). For gel source data, see Supplementary Fig 1. d. Quantification of PLAs between MYC and indicated proteins in DOX (1 µg/ml, 24 h) and PlaB (1 µM, 4 h) treated U2OSMYC-Tet-On cells. Signals are corrected for corresponding single antibody control and presented as fold change to untreated. PLA data were analyzed for the following total number of cells: PLA between MYC and K48 (DMSO: n = 750, PlaB: n = 754), K6 (DMSO: n = 675, PlaB: n = 747), USP28 (DMSO: n = 657, PlaB: n = 554), USP36 (DMSO: n = 799, PlaB: n = 644), HUWE1 (DMSO: n = 343, PlaB: n = 477) analyzed over three independent experiments and are expressed as mean ± S.E.M. Two sided Mann-Whitney U test was used for comparison to untreated cells with following p-values: K48 = 3.3e-11, K6 = 1.8e-3, USP28 = 5.6e-3, USP36 = 1.3e-10, HUWE1 = 5.5e-3. e. Representative images of confocal immunofluorescence of MYC with USP28-MYC and USP36-MYC PLAs within the same cells at indicated treatments. Scale bar: 5 µm. f. Dose response curve of MYC multimer formation in response to BI8626 treatment at indicated concentrations using a four-parameter Weibull function (n = 3). g. Confocal immunofluorescence of MYC and SUMO2/3 in U2OSMYC-Tet-On cells treated as indicated (DOX: 1 µg/ml, 24 h; MG-132: 20 µM, 4 h; ML-792: 1 µM, 24 h). Scale bar: 5 µm (n = 3). In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 9 Role of HUWE1 in MYC multimerization.
a. Representative MS spectrum visualizing MYC ubiquitylation at K148 and K389 in diGLY experiments performed upon treatment with PlaB (2 h, 1 µM), CBL0137 (2 h, 5 µM) or HU (4 h, 5 mM) (n = 3). b. Immunofluorescence pictures of U2OS cells expressing MYCK-R148KR389K. Scale bar: 5 µm (n = 3). c. Fiber assay assessing unperturbed DNA replication (T1) in U2OSMYC-AID expressing either WT, K-less or no MYC. Mean of n = 118 fibers (MYC OFF), n = 127 (WT MYC) and n = 110 (K-less MYC) from one representative of three experiments with similar results is shown ± S.E.M. p-value: Mann-Whitney U test, two-sided. d. BLISS assays in U20SMYC-AID cells reconstituted with MYCK-R148KR389K. MYC OFF samples are the same as shown in Fig. 4f (n = 2). e. Box plot showing quantification of spike normalized reads from EXOSC10 CUT&RUN in U2OSMYC-Tet-On cells on 845 genes, which show increased antisense transcription in absence of MYC. Cells were treated as indicated (MG-132: 20 µM, 4 h; BI8626: 10 µM, 4 h). Box plots are defined as in Fig. 3g. Mann Whitney U test, one-sided: ns, not significant; ****p-value < 0.0001. f. Fiber assay assessing fork degradation (T2) upon treatment with BI8626 (20 µM, 4 h) and Mirin (50 µM, 4 h) in KPC and U2OS cells. Mean of n = 177 (CTL), n = 126 (BI8626), n = 148 (CTL+Mirin) and n = 151 (BI6826+Mirin) fibers from one representative of three experiments with similar results is shown ± S.E.M. for KPC cells (left). Mean of n = 136 (CLT) and n = 72 (BI8626) fibers ± S.E.M. in U2OS cells shown from one representative of three experiments with similar results (right). p-value: Mann-Whitney U test, two sided. g. Fiber assay assessing fork restart (T3; see Extended Data Fig. 7g) upon treatment with BI8626 (20 µM, 4 h) in KPC (left) and U2OS (right) cells. Mean ± S.D. of three independent biological replicates is shown with each replicate quantifying more than 500 fibers per condition (CTL: 505, 506, 501; BI8626: 502, 507, 523) in KPC and more than 400 fibers (CTL: 408, 407, 432; BI8626: 426, 456, 495) in U2OS cells. p-value: unpaired t-test, two sided. In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Extended Data Fig. 10 Control of checkpoint responses by MYC multimers.
a. Immunoblot assessing phosphorylation state or RPA32 at S4/S8 of U2OSMYC-Tet-On cells upon treatment with DOX (1 µg/ml, 24 h), HU (3.5 mM, 8 h) and BI8626 (10 µM, 8.5 h). Chromatin-bound proteins shown with histone H3 as loading control (n = 3). For gel source data, see Supplementary Fig 1. b. Spike normalized reads from pRPAS4/S8 CUT&RUN experiment centered around 4,252 common peaks called around promoters in U2OSMYC-Tet-On cells treated with DOX (1 µg/ml, 24 h), HU (3.5 mM, 8 h) and BI8626 (10 µM, 8.5 h) (n = 1). c. Box plots documenting levels of pS33RPA (Left) and pS4/S8RPA (right) in U2OSMYC-Tet-On cells treated as indicated (DOX: 1 µg/ml, 24 h; HU: 5 mM, 4 h; MG-132: 20 µM, 4 h) Box plots are defined as in Fig. 3g. Data were analyzed from n = 946 cells per condition analyzed over three biological independent experiments. d. Model summarizing our findings. In above panels, unless indicated otherwise, n indicates the number of independent biological replicates.
Supplementary information
Supplementary Information
This file contains Supplementary Fig. 1 Tables 1, 3 and 4 and references.
Supplementary Table 2
Table listing results from APEX2–MYC quantitative mass spectrometry.
Supplementary Video 1
3D-reconstruction of MYC multimers based on Structured Illumination Microscopy.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Solvie, D., Baluapuri, A., Uhl, L. et al. MYC multimers shield stalled replication forks from RNA polymerase. Nature 612, 148–155 (2022). https://doi.org/10.1038/s41586-022-05469-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05469-4
This article is cited by
-
Transcriptional condensates: a blessing or a curse for gene regulation?
Communications Biology (2024)
-
PAF1c links S-phase progression to immune evasion and MYC function in pancreatic carcinoma
Nature Communications (2024)
-
A Machine Learning Computational Framework Develops a Multiple Programmed Cell Death Index for Improving Clinical Outcomes in Bladder Cancer
Biochemical Genetics (2024)
-
Synergistic effect of adavosertib and fimepinostat on acute myeloid leukemia cells by enhancing the induction of DNA damage
Investigational New Drugs (2024)
-
MYC up-regulation confers vulnerability to dual inhibition of CDK12 and CDK13 in high-risk Group 3 medulloblastoma
Journal of Experimental & Clinical Cancer Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.