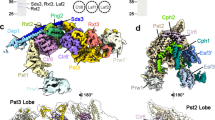
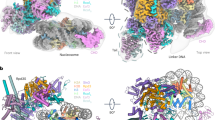
Abstract
Deoxyribonucleic acid in eukaryotes wraps around the histone octamer to form nucleosomes1, the fundamental unit of chromatin. The N termini of histone H4 interact with nearby nucleosomes and play an important role in the formation of high-order chromatin structure and heterochromatin silencing2,3,4. NuA4 in yeast and its homologue Tip60 complex in mammalian cells are the key enzymes that catalyse H4 acetylation, which in turn regulates chromatin packaging and function in transcription activation and DNA repair5,6,7,8,9,10. Here we report the cryo-electron microscopy structure of NuA4 from Saccharomyces cerevisiae bound to the nucleosome. NuA4 comprises two major modules: the catalytic histone acetyltransferase (HAT) module and the transcription activator-binding (TRA) module. The nucleosome is mainly bound by the HAT module and is positioned close to a polybasic surface of the TRA module, which is important for the optimal activity of NuA4. The nucleosomal linker DNA carrying the upstream activation sequence is oriented towards the conserved, transcription activator-binding surface of the Tra1 subunit, which suggests a potential mechanism of NuA4 to act as a transcription co-activator. The HAT module recognizes the disk face of the nucleosome through the H2A–H2B acidic patch and nucleosomal DNA, projecting the catalytic pocket of Esa1 to the N-terminal tail of H4 and supporting its function in selective acetylation of H4. Together, our findings illustrate how NuA4 is assembled and provide mechanistic insights into nucleosome recognition and transcription co-activation by a HAT.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Kayne, P. S. et al. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55, 27–39 (1988).
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T. J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Allard, S. et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18, 5108–5119 (1999).
Bird, A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415 (2002).
Babiarz, J. E., Halley, J. E. & Rine, J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20, 700–710 (2006).
Durant, M. & Pugh, B. F. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 27, 5327–5335 (2007).
Bruzzone, M. J., Grunberg, S., Kubik, S., Zentner, G. E. & Shore, D. Distinct patterns of histone acetyltransferase and mediator deployment at yeast protein-coding genes. Genes Dev. 32, 1252–1265 (2018).
Dhar, S., Gursoy-Yuzugullu, O., Parasuram, R. & Price, B. D. The tale of a tail: histone H4 acetylation and the repair of DNA breaks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160284 (2017).
Doyon, Y. & Cote, J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14, 147–154 (2004).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 8, 284–295 (2007).
Cheung, A. C. M. & Diaz-Santin, L. M. Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes. Transcription 10, 37–43 (2019).
Boudreault, A. A. et al. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17, 1415–1428 (2003).
Xu, P. et al. The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol. Cell 63, 965–975 (2016).
Chittuluru, J. R. et al. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat. Struct. Mol. Biol. 18, 1196–1203 (2011).
Wang, X., Ahmad, S., Zhang, Z., Cote, J. & Cai, G. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat. Commun. 9, 1147 (2018).
Setiaputra, D. et al. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multimodularity. Mol. Cell. Biol. 38, e00570-17 (2018).
Wang, H. et al. Structure of the transcription coactivator SAGA. Nature 577, 717–720 (2020).
Wu, J., Xie, N., Wu, Z., Zhang, Y. & Zheng, Y. G. Bisubstrate inhibitors of the MYST HATs Esa1 and Tip60. Bioorg. Med. Chem. 17, 1381–1386 (2009).
Yuan, H. et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 31, 58–70 (2012).
Sun, B. et al. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J. Biol. Chem. 283, 36504–36512 (2008).
Xu, C., Cui, G., Botuyan, M. V. & Mer, G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16, 1740–1750 (2008).
Rossetto, D. et al. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 33, 1397–1415 (2014).
Lin, L., Chamberlain, L., Zhu, L. J. & Green, M. R. Analysis of Gal4-directed transcription activation using Tra1 mutants selectively defective for interaction with Gal4. Proc. Natl Acad. Sci. USA 109, 1997–2002 (2012).
Knutson, B. A. & Hahn, S. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 31, 818–831 (2011).
Brown, C. E. et al. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337 (2001).
Elias-Villalobos, A., Toullec, D., Faux, C., Seveno, M. & Helmlinger, D. Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes in yeast. Nat. Commun. 10, 5237 (2019).
Auger, A. et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 28, 2257–2270 (2008).
Bittner, C. B., Zeisig, D. T., Zeisig, B. B. & Slany, R. K. Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p. Eukaryot. Cell 3, 976–983 (2004).
Klein, B. J. et al. Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain. Nucleic Acids Res. 46, 421–430 (2018).
Mitchell, L. et al. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 28, 2244–2256 (2008).
Downs, J. A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004).
Turegun, B., Kast, D. J. & Dominguez, R. Subunit Rtt102 controls the conformation of the Arp7/9 heterodimer and its interactions with nucleotide and the catalytic subunit of SWI/SNF remodelers. J. Biol. Chem. 288, 35758–35768 (2013).
Schubert, H. L. et al. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc. Natl Acad. Sci. USA 110, 3345–3350 (2013).
Huang, J. & Tan, S. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell. Biol. 33, 159–169 (2013).
Barbera, A. J. et al. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 (2006).
Steunou, A. L. et al. Combined action of histone reader modules regulates NuA4 local acetyltransferase function but not its recruitment on the genome. Mol. Cell. Biol. 36, 2768–2781 (2016).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–350 (2016).
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Stark, H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Dawson, P. E., Muir, T. W., Clark-Lewis, I. & Kent, S. B. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Li, Y. T. et al. A semisynthetic Atg3 reveals that acetylation promotes Atg3 membrane binding and Atg8 lipidation. Nat. Commun. 8, 14846 (2017).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Ludtke, S. J. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673, 157–173 (2010).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Nakane, T., Kimanius, D., Lindahl, E. & Scheres, S. H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018).
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015).
Diaz-Santin, L. M., Lukoyanova, N., Aciyan, E. & Cheung, A. C. Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution. eLife 6, e28384 (2017).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Ye, Y. et al. Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019).
Graham, M., Combe, C. W., Kolbowski, L. & Rappsilber, J. xiView: a common platform for the downstream analysis of crosslinking mass spectrometry data. Preprint at bioRxiv https://doi.org/10.1101/561829 (2019).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank S.-C. Cheng (Institute of Molecular Biology, Academia Sinica) for providing yeast strain BJ2168 and the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for use of the cryo-EM facility. This work was supported by the National Key Research and Development Program (no. 2019YFA0508902 to Z.C), the National Natural Science Foundation of China (nos. 32130016 and 31825016 to Z.C), Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology and Tsinghua-Peking Joint Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
K.Q. prepared samples and performed biochemical analysis. K.Q. and K.C. built the atomic model. K.C., K.Q., H.W. and X.L. performed EM analysis. Z.C. wrote the manuscript with help from all authors. Z.C. directed and supervised all of the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Biochemistry of NuA4 and the crosslinking data detected by CL-MS.
(a) Preparation of endogenous NuA4 complex from S.cerevisiae. SDS-PAGE gels were showed with Coomassie stain. More sample of the WT complex was loaded to show the weak bands on the right panel. Over 10 batches of the samples were prepared, and the representative two are shown here. (b) Relative HAT activity of NuA4 towards the nucleosomes in the absence and presence of Gal4-VP16. The Gal4 binding site (UAS) was placed at different positions of the nucleosomes (UAS-0N20, UAS-10N20 and UAS-20N20), and the unrelated sequence was used as a control (con-20N20). Acetylation of H4 was detected by Western blotting. A presentative gel was shown on the top, and quantification at the bottom. The activities were normalized to the ones in the absence of Gal4-VP16. Two technical replicates were performed. (c) Overview of the cross-linking data. Circular plot of high confidence lysine-lysine inter-subunit crosslinks obtained by mass spectrometry for the NuA4-NCP complex. (d) Validated cross-links mapped onto the NuA4-NCP structure. Blue lines, the intra-chain cross-links with cross-linked sites within the 30 Å distance permitted by BS3; green lines, the inter-chain cross-links, whereas red lines depict cross-link over more than 30 Å. Bottom graph displays the distribution of cross-link distances. A vertical line indicates a cutoff at 30 Å, the distance considered reasonable for BS3 crosslinks. (e) Relative HAT activity of an independent batch of WT and DM mutant NuA4 different from that of Fig. 2f. Four technical replicates were performed.
Extended Data Fig. 2 Cryo-EM analysis of the NuA4-NCP complex in the absence and presence of Gal4-VP16.
(a) Representative negative stain images (top panel), cryo-EM images (middle panel), and 2D classification (bottom panel) of NuA4+NCP. (b) Flowcharts of the cryo-EM data processing for the dataset of NuA4+NCP. (c) Representative negative stain images (top panel), cryo-EM images (middle panel), and 2D classification (bottom panel) of NuA4+NCP+Gal4-VP16. (d) Flowcharts of the cryo-EM data processing for the dataset of NuA4+NCP+Gal4-VP16.
Extended Data Fig. 3 Chemistry and Mass-spectrometry analyses of the modified histones.
(a) Schematic diagram of the chemical semi-synthesis of histone H4 with CMC modification at Lys16. (b) Mass-spectrometry identification of the CMC-modified H4 (the expected mass is 11092.57 Da). (c) Mass-spectrometry identification of the H3KC36me3-modified H3 (the expected mass is 15299.76 Da).
Extended Data Fig. 4 Cryo-EM analysis of the NuA4-NCP complex in the presence of Gal4-VP16 and histone-modified nucleosome.
(a) Representative negative stain images (left panel), cryo-EM images (middle panel), and 2D classification (right panel) of NuA4+Gal4-VP16 bound to the H4- and H3-modified nucleosome. (b) Flowcharts of the cryo-EM data processing for the dataset of NuA4+Gal4-VP16 bound to the H4- and H3-modified nucleosome. (c-d) Angular distributions of cryo-EM particles in the final round of refinement of the masked dataset and the estimation of the local resolutions of the TRA module (c) and the nucleosome bound HAT module (d). (e) Gold standard Fourier shell correlation (FSC) curves, showing the overall nominal resolutions of 6.7 Å, 3.4 Å, 3.1 Å, 2.8 Å, 2.7 Å and 2.7 Å for the NCP-HAT with longer linker DNA, the nucleosome bound HAT module, TRA module, the NCP bound with arginine anchors (RAs), ARP submodule, head of Tra1 bound with Eaf1/Eaf2/Epl1, respectively.
Extended Data Fig. 5 Local density maps of the NuA4-nucleosome complex.
(a) Local cryo-EM maps of the TRA module. PBL, polybasic loop. (b) Local cryo-EM maps of the HAT module bound to the nucleosome. EM densities of four consecutive base pairs at the central position of the nucleosome are showed on the left.
Extended Data Fig. 6 Multiple sequence alignments of Eaf2-like proteins.
The conserved residues are indicated by “*”. The secondary structural assignments are based on the structure of Eaf2.
Extended Data Fig. 7 Multiple sequence alignments of Epl1-like proteins.
The conserved residues are indicated by “*”. The secondary structural assignments are based on the structure of Epl1.
Extended Data Fig. 8 Multiple sequence alignments of Eaf1-like proteins.
The conserved residues are indicated by “*”. The secondary structural assignments are based on the structure of Eaf1.
Extended Data Fig. 9 Additional structural analysis of the HAT module.
(a) Comparison of the structures of the HAT module bound to the nucleosome (color coded) and the nucleosome-free Piccolo subcomplex (colored grey, PBD code 5J9W)15. The structure of the Esa1 subunit is aligned. (b) Structural alignment of CMC-binding pocket in NuA4 (color coded) and in Esa1 (colored grey, PBD code 3TO6)21. (c) Structural comparison of CMC-binding in NuA4 (color coded) and AcCoA in Piccolo (colored grey, PBD code 5J9W)15.
Supplementary information
Supplementary Fig. 1
Uncropped gels.
Supplementary Table 1
Data of CL–MS.
Supplementary Video 1
Cryo-EM density and model fitting of NuA4 bound to the nucleosome.
Supplementary Video 2
Multi-body refinement analysis of the NuA4–nucleosome complex.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qu, K., Chen, K., Wang, H. et al. Structure of the NuA4 acetyltransferase complex bound to the nucleosome. Nature 610, 569–574 (2022). https://doi.org/10.1038/s41586-022-05303-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05303-x
This article is cited by
-
The NuA4 histone acetyltransferase: variations on a theme of SAGA
Nature Structural & Molecular Biology (2023)
-
Paralog-specific recognition
Nature Chemical Biology (2023)
-
MOF-mediated histone H4 Lysine 16 acetylation governs mitochondrial and ciliary functions by controlling gene promoters
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.