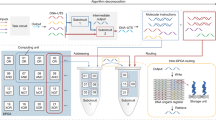
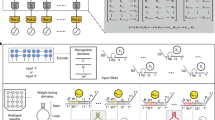
Abstract
Artificial neural networks have revolutionized electronic computing. Similarly, molecular networks with neuromorphic architectures may enable molecular decision-making on a level comparable to gene regulatory networks1,2. Non-enzymatic networks could in principle support neuromorphic architectures, and seminal proofs-of-principle have been reported3,4. However, leakages (that is, the unwanted release of species), as well as issues with sensitivity, speed, preparation and the lack of strong nonlinear responses, make the composition of layers delicate, and molecular classifications equivalent to a multilayer neural network remain elusive (for example, the partitioning of a concentration space into regions that cannot be linearly separated). Here we introduce DNA-encoded enzymatic neurons with tuneable weights and biases, and which are assembled in multilayer architectures to classify nonlinearly separable regions. We first leverage the sharp decision margin of a neuron to compute various majority functions on 10 bits. We then compose neurons into a two-layer network and synthetize a parametric family of rectangular functions on a microRNA input. Finally, we connect neural and logical computations into a hybrid circuit that recursively partitions a concentration plane according to a decision tree in cell-sized droplets. This computational power and extreme miniaturization open avenues to query and manage molecular systems with complex contents, such as liquid biopsies or DNA databases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data are provided with this paper. Source data for the droplet microfluidic experiments are provided within the Supplementary Information.
Code availability
The code is available upon request.
References
Kim, J., Hopfield, J. J. & Winfree, E. Neural network computation by in vitro transcriptional circuits. Adv. Neural Inf. Process. Syst. 17, 681–688 (2004).
Genot, A. J., Fujii, T. & Rondelez, Y. Scaling down DNA circuits with competitive neural networks. J. R. Soc. Interface 10, 20130212 (2013).
Cherry, K. M. & Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 559, 370–376 (2018).
Lopez, R., Wang, R. & Seelig, G. A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 10, 746–754 (2018).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Genot, A. J., Bath, J. & Turberfield, A. J. Reversible logic circuits made of DNA. J. Am. Chem. Soc. 133, 20080–20083 (2011).
Wang, F. et al. Implementing digital computing with DNA-based switching circuits. Nat. Commun. 11, 121 (2020).
Woods, D. et al. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature 567, 366–372 (2019).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Song, T. et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 14, 1075–1081 (2019).
Kim, J., White, K. S. & Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2, 68 (2006).
Montagne, K., Gines, G., Fujii, T. & Rondelez, Y. Boosting functionality of synthetic DNA circuits with tailored deactivation. Nat. Commun. 7, 13474 (2016).
Meijer, L. H. H. et al. Hierarchical control of enzymatic actuators using DNA-based switchable memories. Nat. Commun. 8, 1117 (2017).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Franco, E. et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA 108, E784–E793 (2011).
Kim, J. & Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011).
Fujii, T. & Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 7, 27–34 (2013).
Gines, G. et al. Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 12, 351–359 (2017).
Padirac, A., Fujii, T., Estévez-Torres, A. & Rondelez, Y. Spatial waves in synthetic biochemical networks. J. Am. Chem. Soc. 135, 14586–14592 (2013).
Zambrano, A., Zadorin, A. S., Rondelez, Y., Estévez-Torres, A. & Galas, J.-C. Pursuit-and-evasion reaction-diffusion waves in microreactors with tailored geometry. J. Phys. Chem. B 119, 5349–5355 (2015).
Zadorin, A. S. et al. Synthesis and materialization of a reaction–diffusion French flag pattern. Nat. Chem. 9, 990–996 (2017).
Dupin, A. & Simmel, F. C. Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem. 11, 32–39 (2019).
Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 65, 386–408 (1958).
Slaby, O. et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72, 397–402 (2007).
Genot, A. J. et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 8, 760–767 (2016).
Lobato-Dauzier, N. et al. Silicon chambers for enhanced-imaging of droplet arrays in a graded temperature field. In microTAS 2019 (Chemical and Biological Microsystems Society, 2019).
Mangan, S. & Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003).
Su, J.-L., Chen, P.-S., Johansson, G. & Kuo, M.-L. Function and regulation of let-7 family microRNAs. MicroRNA Shariqah United Arab Emir. 1, 34–39 (2012).
Goldbeter, A. & Koshland, D. E. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl Acad. Sci. USA 78, 6840–6844 (1981).
Vasic, M., Chalk, C., Khurshid, S. & Soloveichik, D. Deep molecular programming: A natural implementation of binary-weight ReLU neural networks. In International Conference on Machine Learning (eds Daumé, H. III & Singh, A.) 9701–9711 (PMLR, 2020).
Hao, Y., Li, Q., Fan, C. & Wang, F. Data storage based on DNA. Small Struct. 2, 2000046 (2021).
Gines, G. et al. Isothermal digital detection of microRNAs using background-free molecular circuit. Sci. Adv. 6, eaay5952 (2020).
McDermott, A. M. et al. Identification and validation of oncologic miRNA biomarkers for Luminal A-like breast cancer. PLoS ONE 9, e87032 (2014).
Lancashire, L. J. et al. A validated gene expression profile for detecting clinical outcome in breast cancer using artificial neural networks. Breast Cancer Res. Treat. 120, 83–93 (2010).
Yamagata, A., Masui, R., Kakuta, Y., Kuramitsu, S. & Fukuyama, K. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Res. 29, 4617–4624 (2001).
Van Der Hofstadt, M., Galas, J.-C. & Estevez-Torres, A. Spatiotemporal patterning of living cells with extracellular DNA programs. ACS Nano 15, 1741–1752 (2021).
Baccouche, A. et al. Massively parallel and multiparameter titration of biochemical assays with droplet microfluidics. Nat. Protoc. 12, 1912–1932 (2017).
Deteix, R. et al. Droplet-based investigation of a biochemical bistable circuit for sensitive and noise-free detection of nucleic acid. In microTAS 2019 (Chemical and Biological Microsystems Society, 2019).
Peng, T. et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8, 14836 (2017).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Acknowledgements
This research was supported by the French ANR (grant SmartGuide to A.J.G.), the Japanese JSPS (DC1 fellowship 18J22815 to S.O., postdoctoral fellowship 17F17796 to A.B. and Core-to-Core Program on Advanced Research Networks to T.F.), the Japanese MEXT (studentship to N.L.D. and R.D.), the ERC (CoG ProFF 647275 to Y.R. and StG MoP-MiP 949493 to G.G.) and the CNRS (MITI DNA2 grant to A.J.G. and G.G.). We also acknowledge support from the RENATECH microfabrication network in France for the fabrication of the silicon chambers. A.J.G acknowledges support from the ESPCI (Joliot chair) and G.G. acknowledges support from LIMMS for a travel grant. We thank N. Aubert-Kato and L. Cazenille for discussions.
Author information
Authors and Affiliations
Contributions
S.O., G.G. and N.L-D. performed the experiments. N.L-D. and R.D. designed the silicon chambers and the thermal gradient platform. A.B. provided support with the enzymatic toolbox. T.F., Y.R., G.G. and A.J.G. supervised the research.
Corresponding author
Ethics declarations
Competing interests
T.F., Y.R. and G.G. have filed a patent on the PEN DNA toolbox (patent no. WO2017141067A1).
Peer review
Peer review information
Nature thanks Chunhai Fan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Experimental validation of the basic components of a chemical neuron.
To demonstrate the weighted summation mechanism (a, weight adjustment and b, summation), we used high concentrations of input strands, which allows for the direct visualization of the linear production of α strand (i.e. in absence of the amplification reaction that composes the activation function). The threshold activation function and the possibility to control the bias (c) is estimated by measuring the concentration of α required to trigger the amplification reaction at given drain template (dTα) concentrations. a, Weight adjustment: w1 corresponds to the ratio of X1toα (cT) and X1tof (fT) (10 nM total). The production of α from various concentrations of X1 is directly monitored using 25 nM of rTα. b, Summation of X1 and X2: all samples contain 5 nM X1toα, 5 nM X1tof (w1 = 0.5) and 10 nM X2toα (w2 = 1). The production of α from different concentrations of X1 and X2 is directly monitored using 25 nM of rTα. c, Activation function: the amplification reaction of samples containing various initial concentrations of α is monitored in real-time. The bias (i.e. amplification threshold, noted t) is tuned according to the concentration of dTα.
Extended Data Fig. 2 Thermal setup for droplet incubation and imaging.
a, After generation, an emulsion is spread into a monolayer of droplets inside a silicon chamber. Silicon offers ideal conditions for imaging and incubation: high thermal conductivity, mechanical rigidity and optical reflectivity. The inset shows a monolayer of droplets imaged by fluorescence. Scale bar = 500 μm. b, Bottom view of the setup. The chamber is fixed against a copper plate by capillarity by sandwiching a drop of mineral oil between the plate and the chamber. Two Peltier elements, separated by ~ 7.5 cm, impose a thermal gradient across the copper plate. Pt100 sensors report the local temperature near each Peltier element to a Peltier controller. The whole setup is encased into a 3D printed frame that is fitted into the microscope stage. c, Side view of the setup. Heat is extracted from the Peltier elements with CPU cooling fans.
Extended Data Fig. 3 Microfluidic workflow for measuring the dependence of the separatrix on the concentration of reagents.
a, We determined the equation of the separatrix in the (X1,X2) plane for 3 conditions in a multiplexed experiment. We first performed 3 rounds of droplet generation. During each round, we scanned the (X1,X2) plane by mixing 3 tubes (which all derive from the same master mix) and collected the emulsion in a separate tube. Between each round, we changed the set of 3 tubes - thus changing their common mastermix. This allowed us to vary the concentrations of converter templates or drain templates in the master mix. After generation, we simultaneously imaged the three subpopulations together. To that end, we sequentially and gently filled a chamber with each population: the subpopulations remained spatially separated. We then incubated and imaged the chamber. During image processing, we separated the subpopulation by selecting 3 distinct regions (shown as red boxes in the right picture).
Extended Data Fig. 4 Design and kinetics of the linear classifier.
a, Full architecture of the linear classifier for one input X1. The classifier comprises three templates: a converter template (which produces α when bound to its input X1), a autocatalytic template (which autocatalytically replicates α), and a drain template (which deactivates α). The strand α is continuously degraded by the exonuclease. b, Production and removal curves showing the rate of production of α by the autocatalytic template (left), and the rate of removal of α by the drain template and the exonuclease (middle). Inactivation by drain template is fast but quickly saturated, whereas degradation by the exonuclease is slow but linear. This interplay creates a kink in the removal curve, resulting in the existence of 3 intersection points between the production curve and the removal curve. The upper and lower intersection points are the two stable steady states (OFF and ON), whereas the middle point is an unstable steady state, a threshold which controls the crossover between autocatalytic production and removal. If α is over the threshold, the drain is saturated, production overcomes the removal by the exonuclease and the drain, and α is amplified up to the ON state. Otherwise, α is removed down to the OFF state. The existence of an unstable steady state is controlled by the shape of production and removal curves, which must intersect at three points. The shape of the removal curve is controlled by the concentrations of drain templates, exonuclease and polymerase (for the inactivation step in the drain). The shape of the production curve is controlled by the concentration of autocatalytic templates, polymerase and nickase. c, Microfluidic mapping of the dependence of the bias on X1 to the drain. We prepared droplets with varying concentrations of drain for α and input X1. We incubated the droplets in a temperature gradient, and imaged their content after 6 h. The top plots show the fluorescence of droplets in the space (drain, X1), the colour indicating the level of α. The bottom plots show the fluorescence of droplets in the space (temperature, X1). The red lines are linear fit of the boundary, with equation indicated above each plot. The concentration are in nM and the temperature in Celsius.
Extended Data Fig. 5 Enzymatic classifiers have stronger nonlinearities and higher sensitivities than a state-of-the-art nonenzymatic classifier.
a, Steady-state fluorescence of a reported negative-weighted nonenzymatic linear classifier4. The fitted Hill equation cn/(cn+xn) is shown in plain, the interpolation from the 8 points along the diagonal of Fig. 3d of ref. 4 is shown in dashed. b, Steady-state fluorescence of our negative-weighted enzymatic linear classifier (Fig. 2d, left), and its fit by a Hill equation cn/(cn+xn). c, Steady-state fluorescence of our positive-weighted enzymatic linear classifier (Fig. 2c, middle) and its fit by a Hill equation xn/(cn+xn). Hill equations were fitted with a prefactor to improve the goodness of fit.
Extended Data Fig. 6 Majority voting weight adjustment.
a-b. Trigger production by 10 converter templates at 1 nM concentration (a) or adjusted proportionally to the production rate, arbitrarily choosing X3 as reference (b). c. Comparison of the production rate from panel a. and b. At constant cT concentration, we observed large rate discrepancies depending on the input sequence (mean production rate = 39 pM ± 18 pM/min). A factor of 4.6 was computed between the fastest and the slowest converter template. Balancing the cT concentration reduces the coefficient of variation from 47 % to 14 % (29 +/−4 pM/min). d-e. Majority voting with balanced cT concentrations. In a first attempt, we perform a majority voting experiment on 28 samples. Concentrations of the 1X cT bundle were 0.8, 0.7, 1.2, 1.9, 3, 0.6, 1.2, 0.5, 1.4, 1.4 nM, respectively from X1 to X10. Enzyme concentrations were set to 70 u/mL Vent(exo-), 300 u/mL Nb.BsmI and 23 nM of ttRecJ. d. Amplification curves for the 28 samples spiked with 0 to 10 inputs (5 nM each, various combinations). e. Bar chart of the amplification times (Cq). As expected, Cq are negatively correlated to the number of spiked inputs (the more inputs, the faster the amplification). Interestingly, all ten inputs exert a consistent activation force on the switch, triggering the amplification between 112 and 148 min when spiked individually (123 ± 11 min on average, hence a coefficient of variation of less than 10%). In these conditions, all samples amplify within 150 min (except for the negative control, NC), suggesting that the production rate for each input is too high to unambiguously classify samples with less than 5 inputs from samples with more than 5 inputs. f. Effect of the cT bundle concentration on sample classification. To further reduce the weight of all inputs, we decreased the concentration of all converter templates from 1 X to 0.04 X. For the lowest concentration of cT bundle, no amplification is observed within 1000 min for all samples (from 0 to 6 inputs). 1X cT bundle results reproducibly in the amplification of all samples (except the negative control, NC), with a poor discrimination between 4, 5 and 6 inputs samples. Interestingly, we observed a sharp threshold between ≤ 2 and > 2 inputs with a 3-fold dilution of the cT bundle concentration. This demonstrates that the amplification threshold can be tuned finely by adjusting the production rate of all inputs, allowing to set an arbitrary number of input voters to return a positive answer. Finally, 0.15 X of cT bundle allows us to compute a majority voting algorithm, set a clear discrimination between 4 and 6 inputs.
Extended Data Fig. 7 Operation of the NOR gate.
a, A NOR gate is formed by hybridizing a template strand (accepting α or β as primers) with a molecular beacon γ. In the absence of α and β, the fluorescence of the molecular beacon is high because its dye and quencher are far apart. This codes for the ON state of the gate. If α or β is present, it induces polymerase-mediated displacement of the molecular beacon from the template, resulting in a low level of fluorescence. b, Verification of the assembly of the beacon with the template. The fluorescence of the beacon is monitored as the solution is cooled in a thermocycler, with and without template. The presence of the template induces a large fluorescence increase, due to the hybridization of the beacon. c, A two-layer network with the NOR gate as its output (left) and its truth table (middle) The network consists of two linear classifiers (α and β), each accepting a distinct input (X1 or X2). The correct operation of the network is verified by fluorescence: presence of either input drives the polymerization of the gate and the displacement of the beacon, leading to a drop in fluorescence.
Extended Data Fig. 8 Thermal dependence of the space partitioning network (Fig. 4).
Droplets with the network and varying inputs were incubated in a graded temperature field to reveal how the quality of classification varies with temperature. At low temperature (~43 °C), α and β coexist, as indicated by the yellow region. When temperature increases, the yellow region shrinks and disappears, indicating that α and β do not coexist.
Supplementary information
Supplementary Information
Supplementary materials and methods, mathematical model, additional experiments and discussions.
Supplementary Methods
Experimental conditions.
Supplementary Table 1
Nucleic acid sequences.
Supplementary Data
Supplementary data and source code used to plot Figs. 1–4.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Okumura, S., Gines, G., Lobato-Dauzier, N. et al. Nonlinear decision-making with enzymatic neural networks. Nature 610, 496–501 (2022). https://doi.org/10.1038/s41586-022-05218-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05218-7
This article is cited by
-
Pattern recognition in the nucleation kinetics of non-equilibrium self-assembly
Nature (2024)
-
A genetic circuit on a single DNA molecule as an autonomous dissipative nanodevice
Nature Communications (2024)
-
A signal transmission strategy driven by gap-regulated exonuclease hydrolysis for hierarchical molecular networks
Communications Biology (2024)
-
Functional analysis of single enzymes combining programmable molecular circuits with droplet-based microfluidics
Nature Nanotechnology (2024)
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.