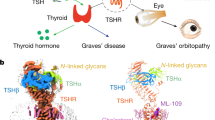
Abstract
Thyroid hormones are vital in metabolism, growth and development1. Thyroid hormone synthesis is controlled by thyrotropin (TSH), which acts at the thyrotropin receptor (TSHR)2. In patients with Graves’ disease, autoantibodies that activate the TSHR pathologically increase thyroid hormone activity3. How autoantibodies mimic thyrotropin function remains unclear. Here we determined cryo-electron microscopy structures of active and inactive TSHR. In inactive TSHR, the extracellular domain lies close to the membrane bilayer. Thyrotropin selects an upright orientation of the extracellular domain owing to steric clashes between a conserved hormone glycan and the membrane bilayer. An activating autoantibody from a patient with Graves’ disease selects a similar upright orientation of the extracellular domain. Reorientation of the extracellular domain transduces a conformational change in the seven-transmembrane-segment domain via a conserved hinge domain, a tethered peptide agonist and a phospholipid that binds within the seven-transmembrane-segment domain. Rotation of the TSHR extracellular domain relative to the membrane bilayer is sufficient for receptor activation, revealing a shared mechanism for other glycoprotein hormone receptors that may also extend to other G-protein-coupled receptors with large extracellular domains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Coordinates for TSH-bound TSHR–Gs, TR1402-bound TSHR–Gs, M22-bound TSHR–Gs and CS-17-bound TSHR have been deposited in the PDB under accession codes 7T9I, 7UTZ, 7T9N and 7T9M, respectively. Sharpened and unsharpened cryo-EM density maps for TSH-bound TSHR–Gs (composite), TR1402-bound TSHR–Gs (composite), M22-bound TSHR–Gs (composite), CS-17-bound TSHR, and Org 2274179-0-bound TSHR have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25758, EMD-26795, EMD-25763, EMD-25762 and EMD-27640, respectively. Sharpened maps, unsharpened maps, half-maps and masks for each composite map component (ligand-bound ECD or 7TM–G protein), for TSH-bound TSHR–Gs, TR1402-bound TSHR–Gs and M22-bound TSHR-Gs have been deposited in the Electron Microscopy Data Bank under accession codes EMD-27649 (TSH-bound ECD), EMD-27650 (TSH-bound 7TM–Gs), EMD-27647 (TR1402-bound ECD), EMD-27648 (TR1402 7TM–Gs), EMD-27651 (M22-bound ECD) and EMD-27652 (M22-bound 7TM–Gs). Final particle stacks and .star files for TSH-bound TSHR–Gs, TR1402-bound TSHR–Gs and M22-bound TSHR–Gs and CS-17-bound TSHR-containing particle shift and pose assignments have been uploaded to the Electron Microscopy Public Image Archive under the accession codes EMPIAR-11120. Structure files for OPM-based modelling of inactive and active TSHR are available in the Supplementary Information.
References
Oppenheimer, J. H. & Samuels, H. H. Molecular Basis of Thyroid Hormone Action (Academic Press, 1983).
Vassart, G. & Dumont, J. E. The thyrotropin receptor and the regulation of thyrocyte function and growth. Endocr. Rev. 13, 596–611 (1992).
Smith, T. J. & Hegedüs, L. Graves’ disease. N. Engl. J. Med. 375, 1552–1565 (2016).
Fekete, C. & Lechan, R. M. Central regulation of hypothalamic–pituitary–thyroid axis under physiological and pathophysiological conditions. Endocr. Rev. 35, 159–194 (2014).
Laurent, E., Mockel, J., Van Sande, J., Graff, I. & Dumont, J. E. Dual activation by thyrotropin of the phospholipase C and cyclic AMP cascades in human thyroid. Mol. Cell. Endocrinol. 52, 273–278 (1987).
Taylor, P. N. et al. Global epidemiology of hyperthyroidism and hypothyroidism. Nat. Rev. Endocrinol. 14, 301–316 (2018).
Zimmermann, M. B. & Boelaert, K. Iodine deficiency and thyroid disorders. Lancet Diabetes Endocrinol. 3, 286–295 (2015).
Mincer, D. L. & Jialal, I. Hashimoto Thyroiditis. in StatPearls (StatPearls Publishing, 2021); https://www.ncbi.nlm.nih.gov/books/NBK459262/
Lane, L. C., Cheetham, T. D., Perros, P. & Pearce, S. H. S. New therapeutic horizons for Graves’ hyperthyroidism. Endocr. Rev. 41, 873–884 (2020).
Flack, M. R., Froehlich, J., Bennet, A. P., Anasti, J. & Nisula, B. C. Site-directed mutagenesis defines the individual roles of the glycosylation sites on follicle-stimulating hormone. J. Biol. Chem. 269, 14015–14020 (1994).
Matzuk, M. M., Keene, J. L. & Boime, I. Site specificity of the chorionic gonadotropin N-linked oligosaccharides in signal transduction. J. Biol. Chem. 264, 2409–2414 (1989).
Grossmann, M., Weintraub, B. D. & Szkudlinski, M. W. Novel insights into the molecular mechanisms of human thyrotropin action: structural, physiological, and therapeutic implications for the glycoprotein hormone family. Endocr. Rev. 18, 476–501 (1997).
Jiang, X. et al. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc. Natl Acad. Sci. USA 109, 12491–12496 (2012).
Duan, J. et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature 598, 688–692 (2021).
Chazenbalk, G. D. et al. Evidence that the thyrotropin receptor ectodomain contains not one, but two, cleavage sites. Endocrinology 138, 2893–2899 (1997).
Chen, C.-R., Salazar, L. M., McLachlan, S. M. & Rapoport, B. Deleting the redundant TSH receptor C-peptide region permits generation of the conformationally intact extracellular domain by insect cells. Endocrinology 156, 2732–2738 (2015).
Nehmé, R. et al. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS ONE 12, e0175642 (2017).
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Szkudlinski, M. W., Teh, N. G., Grossmann, M., Tropea, J. E. & Weintraub, B. D. Engineering human glycoprotein hormone superactive analogues. Nat. Biotechnol. 14, 1257–1263 (1996).
Reinfelder, J. et al. Effects of recombinant human thyroid-stimulating hormone superagonists on thyroidal uptake of 18F-fluorodeoxyglucose and radioiodide. Thyroid 21, 783–792 (2011).
Ulloa-Aguirre, A., Timossi, C., Damián-Matsumura, P. & Dias, J. A. Role of glycosylation in function of follicle-stimulating hormone. Endocrine 11, 205–215 (1999).
Fan, Q. R. & Hendrickson, W. A. Structure of human follicle-stimulating hormone in complex with its receptor. Nature 433, 269–277 (2005).
Costagliola, S. et al. Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. EMBO J. 21, 504 (2002).
Kosugi, S., Ban, T., Akamizu, T. & Kohn, L. D. Site-directed mutagenesis of a portion of the extracellular domain of the rat thyrotropin receptor important in autoimmune thyroid disease and nonhomologous with gonadotropin receptors. Relationship of functional and immunogenic domains. J. Biol. Chem. 266, 19413–19418 (1991).
Caltabiano, G. et al. The specificity of binding of glycoprotein hormones to their receptors. Cell. Mol. Life Sci. 65, 2484–2492 (2008).
Moyle, W. R. et al. Co-evolution of ligand-receptor pairs. Nature 368, 251–255 (1994).
Lapthorn, A. J. et al. Crystal structure of human chorionic gonadotropin. Nature 369, 455–461 (1994).
Wu, H., Lustbader, J. W., Liu, Y., Canfield, R. E. & Hendrickson, W. A. Structure of human chorionic gonadotropin at 2.6 Å resolution from MAD analysis of the selenomethionyl protein. Structure 2, 545–558 (1994).
Grossmann, M. et al. Substitution of the seat-belt region of the thyroid-stimulating hormone (TSH) β-subunit with the corresponding regions of choriogonadotropin or follitropin confers luteotropic but not follitropic activity to chimeric TSH. J. Biol. Chem. 272, 15532–15540 (1997).
Dias, J. A., Zhang, Y. & Liu, X. Receptor binding and functional properties of chimeric human follitropin prepared by an exchange between a small hydrophilic intercysteine loop of human follitropin and human lutropin. J. Biol. Chem. 269, 25289–25294 (1994).
Vischer, H. F., Granneman, J. C. M. & Bogerd, J. Opposite contribution of two ligand-selective determinants in the N-terminal hormone-binding exodomain of human gonadotropin receptors. Mol. Endocrinol. 17, 1972–1981 (2003).
Chen, F., Wang, Y. & Puett, D. Role of the invariant aspartic acid 99 of human choriogonadotropin β in receptor binding and biological activity. J. Biol. Chem. 266, 19357–19361 (1991).
Smits, G. et al. Glycoprotein hormone receptors: determinants in leucine-rich repeats responsible for ligand specificity. EMBO J. 22, 2692–2703 (2003).
van Koppen, C. J. et al. Mechanism of action of a nanomolar potent, allosteric antagonist of the thyroid-stimulating hormone receptor. Br. J. Pharmacol. 165, 2314–2324 (2012).
Van Sande, J. et al. In Chinese hamster ovary K1 cells dog and human thyrotropin receptors activate both the cyclic AMP and the phosphatidylinositol 4,5-bisphosphate cascades in the presence of thyrotropin and the cyclic AMP cascade in its absence. Eur. J. Biochem. 229, 338–343 (1995).
Chen, C.-R., McLachlan, S. M. & Rapoport, B. A monoclonal antibody with thyrotropin (TSH) receptor inverse agonist and TSH antagonist activities binds to the receptor hinge region as well as to the leucine-rich domain. Endocrinology 150, 3401–3408 (2009).
Mueller, S., Jaeschke, H., Günther, R. & Paschke, R. The hinge region: an important receptor component for GPHR function. Trends Endocrinol. Metab. 21, 111–122 (2010).
Mizutori, Y., Chen, C.-R., McLachlan, S. M. & Rapoport, B. The thyrotropin receptor hinge region is not simply a scaffold for the leucine-rich domain but contributes to ligand binding and signal transduction. Mol. Endocrinol. 22, 1171–1182 (2008).
Brüser, A. et al. The activation mechanism of glycoprotein hormone receptors with implications in the cause and therapy of endocrine diseases. J. Biol. Chem. 291, 508 (2016).
Schulze, A. et al. The intramolecular agonist is obligate for activation of glycoprotein hormone receptors. FASEB J. 34, 11243–11256 (2020).
Parma, J. et al. Somatic mutations causing constitutive activity of the thyrotropin receptor are the major cause of hyperfunctioning thyroid adenomas: identification of additional mutations activating both the cyclic adenosine 3′,5′-monophosphate and inositol phosphate-Ca2+ cascades. Mol. Endocrinol. 9, 725–733 (1995).
Vlaeminck-Guillem, V., Ho, S.-C., Rodien, P., Vassart, G. & Costagliola, S. Activation of the cAMP pathway by the TSH receptor involves switching of the ectodomain from a tethered inverse agonist to an agonist. Mol. Endocrinol. 16, 736–746 (2002).
Sanders, J. et al. Human monoclonal thyroid stimulating autoantibody. Lancet 362, 126–128 (2003).
Sanders, J. et al. Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody. Thyroid 17, 395–410 (2007).
Evans, M. et al. Monoclonal autoantibodies to the TSH receptor, one with stimulating activity and one with blocking activity, obtained from the same blood sample. Clin. Endocrinol. 73, 404–412 (2010).
Sanders, P. et al. Crystal structure of the TSH receptor (TSHR) bound to a blocking-type TSHR autoantibody. J. Mol. Endocrinol. 46, 81–99 (2011).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Reily, C., Stewart, T. J., Renfrow, M. B. & Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019).
Erbel, P. J. A., Haseley, S. R., Kamerling, J. P. & Vliegenthart, J. F. G. Studies on the relevance of the glycan at Asn-52 of the α-subunit of human chorionic gonadotropin in the αβ dimer. Biochem. J. 364, 485–495 (2002).
Amr, S. et al. Activities of deglycosylated thyrotropin at the thyroid membrane receptor-adenylate cyclase system. J. Endocrinol. Invest. 8, 537–541 (2014).
Fares, F. A., Levi, F., Reznick, A. Z. & Kraiem, Z. Engineering a potential antagonist of human thyrotropin and thyroid-stimulating antibody. J. Biol. Chem. 276, 4543–4548 (2001).
Feng, X., Müller, T., Mizrachi, D., Fanelli, F. & Segaloff, D. L. An intracellular loop (IL2) residue confers different basal constitutive activities to the human lutropin receptor and human thyrotropin receptor through structural communication between IL2 and helix 6, via helix 3. Endocrinology 149, 1705–1717 (2008).
Zhang, M. et al. Intrinsic differences in the response of the human lutropin receptor versus the human follitropin receptor to activating mutations. J. Biol. Chem. 282, 25527–25539 (2007).
Skiba, M. A. & Kruse, A. C. Autoantibodies as endogenous modulators of GPCR signaling. Trends Pharmacol. Sci. 42, 135–150 (2021).
Girdlestone, C. & Hayward, S. The DynDom3D webserver for the analysis of domain movements in multimeric proteins. J. Comput. Biol. 23, 21–26 (2016).
Sanders, J. et al. Characteristics of a human monoclonal autoantibody to the thyrotropin receptor: sequence structure and function. Thyroid 14, 560–570 (2004).
Mastronarde, D. N. SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183 (2003).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Zhuang, Y. et al. Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor. Cell Res. 31, 593–596 (2021).
Schüttelkopf, A. W. & van Aalten, D. M. F. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Marty, M. T. et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015).
Zhang, L. & Hermans, J. Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996).
Betz, R. Dabble. https://doi.org/10.5281/zenodo.836914 (2017).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Case, D. A. et al. Amber 2021 (Univ. California, San Francisco, 2021).
Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015).
Roe, D. R. & Cheatham, T. E. III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996).
Latorraca, N. R. et al. How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell 183, 1813–1825.e18 (2020).
Asarnow, D., Palovcak, E. & Cheng, Y. UCSF pyem v0. 5. https://doi.org/10.5281/zenodo.3576630 (2019).
Acknowledgements
We thank R. Yan and D. Toso for help in microscope operation and data collection; C. Bracken for mass spectrometry troubleshooting; and H. Shan for sample preparation for lipid mass spectrometry. This work was supported by National Institutes of Health (NIH) grants DP5OD023048 (A.M.), 1R35GM140847 (Y.C.), P30CA014195, R01GM102491 (A.S.), R01GM127359 (R.O.D.), 5R44CA224376 (M.W.S.) and a Human Frontier Science Program Long-Term Fellowship LT000916/2018-L (C.-M.S.). Cryo-EM equipment at UCSF is partially supported by NIH grants S10OD020054 and S10OD021741. Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2), which is supported by the National Institutes of Health Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (U24 GM129541). This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. Some of this work was supported by the Mass Spectrometry Core of the Salk Institute with funding from NIH-NCI CCSG: P30CA014195. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.M. acknowledges support from the Pew Charitable Trusts, the Esther A. & Joseph Klingenstein Fund, the Searle Scholars Program, the Edward Mallinckrodt Jr Foundation, and the Vallee Foundation. Y.C. is an Investigator of Howard Hughes Medical Institute. A.M. is a Chan Zuckerberg Biohub investigator.
Author information
Authors and Affiliations
Contributions
B.F. cloned, expressed, and biochemically optimized the purification of all TSHR constructs for structural studies. B.F. expressed and purified Gβ1γ2, Nb35, M22 Fab, CS-17 IgG and K1-70 IgGs, and performed enzymatic digestion and further purification to form CS-17 and K1-70 Fab fragments. B.F. performed complexing and identified optimal cryo-EM grid preparation procedures, screened samples, and collected 300 keV datasets. B.F. determined high-resolution cryo-EM maps by extensive image processing under the guidance of A.M. and Y.C. B.F. and A.M built and refined models of TSHR complexes. B.F. and C.B.B. generated receptor constructs and determined expression levels by flow cytometry and performed signalling studies and analysed the data. C.B.B. assisted with cloning of TSHR constructs, expression, purification and labelling of CS-17 and M22 Fab fragments, and cloning and generation of baculoviruses for expression of Gβ1γ2. C.-M.S. performed and analysed molecular dynamics simulations under the supervision of R.O.D. I.S. performed initial biochemical optimization of TSHR complexes with M22 Fab and worked with K.Z. to collect negative stain and cryo-EM data. N.H. prepared samples for, performed, and analysed native mass spectrometry experiments and prepared control samples for lipid identification experiments. J.K.D. analysed native human TSH glycosylation by mass spectrometry. Y.M. generated TSHR and M22 Fab expression constructs and performed pilot biochemical purification of TSHR complexes. A.F.M.P. performed and analysed data from the lipid identification experiments with guidance from A.S. M.W.S. led production of highly pure TR1402 agonist. Figures were generated and the manuscript was written by B.F., C.B.B. and A.M., with edits from Y.C., R.O.D., C.-M.S., and M.W.S. and with approval from all authors. The overall project was supervised by Y.C. and A.M.
Corresponding authors
Ethics declarations
Competing interests
A.M. and R.O.D. are consultants for and stockholders in Septerna Inc. Y.C. is a consultant and advisor of Shuimu BioScience Ltd.
Peer review
Peer review information
Nature thanks Marvin Gershengorn, Patrick M. Sexton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Signaling studies for TSHR.
a, b) DNA titrations of TSHR mutants tested in cAMP production assays. Median fluorescence intensity ± SD (n = 3) of anti-FLAG-A647 staining is plotted. Filled in bars represent the DNA concentration used for co-transfection experiments with pGlo cAMP biosensor plasmid. Horizontal red line indicates the mean wild-type (WT) TSHR fluorescence intensity. Flow cytometry experiments were performed in triplicate, with identical gating across all cell lines tested. c) cAMP production of TSHR cysteine mutants comparing the basal level (-) to 100 nM of TSH, M22, or CS-17 for untreated cells (left panel) and cells treated with 500 µM TCEP (right panel). Data points are means ± SD from three biological replicates. d, e) cAMP production curves comparing TSH and M22-mediated activation of WT and C-peptide deleted TSHR constructs. f) cAMP production curves for TSH and TR1402-mediated WT TSHR activation. g) cAMP production curves for TSH-mediated WT and TSHR mutant cell line (Y385F, Y385A) activation. h) cAMP production curves for M22-mediated WT and lipid-displacing TSHR mutant cell line (A644K, A647K) activation. Plotted data points in panels c-g are means of triplicate measurements ± SD from a representative experiment of n = 3 biological replicates.
Extended Data Fig. 2 Cryo-EM data processing for TSH-bound TSHR-Gs complex.
a) Representative image from 15,345 micrographs. Scale bar, 50 nm. b) Selected 2D class averages. c) Processing approach used for reconstruction of TSH-bound TSHR-Gs complex. A local resolution map was calculated from cryoSPARC using masks from indicated local refinement, then visualized with the composite map in the same scale. A viewing distribution plot was generated using scripts from the pyEM software suite83 and visualized in ChimeraX. GS-FSC and Directional FSC (dFSC, shown as purple lines) curves were generated in cryoSPARC and as previously described in Dang, S. et al. Nature 552, 426–429 (2017).
Extended Data Fig. 3 Model map correlation coefficients for cryo-EM structures.
Correlation values for resolved residues in each modeled chain are shown. Low values indicate regions that are poorly resolved, e.g. the 7TM domain of TSHR in CS-17-TSHR complex or the constant domains of the Fab fragments for CS-17 and M22.
Extended Data Fig. 4 Cryo-EM data processing for TR1402-bound TSHR-Gs complex.
a) Representative image from 14,277 micrographs. Scale bar, 50 nm. b) Selected 2D class averages from final reconstruction. c) Processing approach used for reconstruction of TR1402-bound TSHR-Gs complex. A local resolution map was calculated from cryoSPARC using masks from indicated local refinement, then visualized with the composite map in the same scale. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. GS-FSC and dFSC curves were generated in cryoSPARC and as previously described in Dang, S. et al. Nature 552, 426–429 (2017).
Extended Data Fig. 5 Glycosylation status of native human TSH.
a) Cryo-EM density map of TSH-bound TSHR showing resolved glycan density for Asn52. b) Peptide coverage of Asn52 from native human TSH GPHα chain in mass spectrometry experiments. c) Peptide spectra match (PSM) of glycans detected on Asn52 of GPHα chain. d,e) Representative MS/MS spectra showing Asn52 with various length glycans.
Extended Data Fig. 6 Receptor:hormone interaction comparisons across the glycoprotein hormone receptor family.
a) Comparison of TSHR, FSHR (PDB: 4AY9), and LH/CGR ECD:hormone (PDB: 7FIH) interactions shows the comparable concave binding-interface and lateral-ECD contacts made by GPHα and receptor-specific β subunits. b) Alignment of the GPHR ECDs reveals the variability in modeled poses of hinge-hormone contacts observed in prior GPHR:hormone structures. c) The N-terminal hinge-hormone disulfide-linked alpha helix presents a hydrophobic face to the α-L1 and α-L3 loops of the TR1402 α-chain and is stabilized by multiple phenylalanine contacts in the GPHα and TSHβ chains. C-terminal hinge-region α helix hidden for visualization. d) Comparison of the TSHR and LHCGR (PDB: 7FIH) transmembrane pockets highlights the similarity in the DPPC and the allosteric agonist Org43554 binding sites. TM4/5 and ECL2 not shown. e) Lipid:protein molar ratio comparison between the M22:TSHR:Gs complex and a non-GPHR Class A receptor control. Data points represent individual measurements from technical replicates (n = 3 for non-GPHR control and n = 4 for M22:TSHR:Gs complex) of the ratio of pmol of lipid DPPC per pmol of protein. **P = 0.0088; Unpaired two-tailed t test was used to calculate statistical differences in lipid:protein molar ratios.
Extended Data Fig. 7 Cryo-EM data processing for Org 274179-0 bound TSHR.
a) Representative image from 10,003 micrographs. Scale bar, 50 nm. b) Selected 2D class averages generated from curated particles. c) Processing approach used for low resolution reconstruction. Despite starting with a similar or larger sized dataset as for other TSHR samples, the TSHR–Org 274179-0 complex did not yield high resolution reconstruction of the 7TM domain. A low-resolution reconstruction of the TSHR ECD was observed. This suggests potential flexibility between the TSHR 7TM domain and the ECD.
Extended Data Fig. 8 Cryo EM data processing for the CS-17 bound TSHR:Org 274179-0 complex.
a) Representative image from 4,952 micrographs. Scale bar, 50 nm. b) Selected 2D class averages generated from the final reconstruction. c) Processing approach used for reconstruction of the complex. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. Local resolution map generated from non-uniform refinement mask in cryoSPARC. GS-FSC and dFSC curves were generated in cryoSPARC and as previously described in Dang, S. et al. Nature 552, 426–429 (2017).
Extended Data Fig. 9 Comparison of Org 274179-0 bound- and CS-17 bound TSHR EM maps.
a) Org 274179-0 bound-TSHR EM density map (dark) fit into the CS-17-bound TSHR map (colored by TSHR-CS-17 model). b) CS-17-bound TSHR model fit into Org 274179-0 bound-TSHR map, suggesting that inactive state TSHR ECD orientations are similar between CS-17 and Org 274179-0-bound states.
Extended Data Fig. 10 Comparison of TSHR activation with other GPCRs.
a) Ribbon diagram of TSHR in active and inactive conformations. b) Ribbon diagram of LH/CGR in active (PDB:7FIH) and inactive (PDB:7FIJ) conformations. A similar reorientation of the ECD is shared between TSHR and LH/CGR upon activation. c) Comparison of active TSHR to active β2-adrenoceptor (β2AR, PDB:3SN6). d) Comparison of inactive TSHR to inactive β2AR, PDB:5JQH). e) Comparison of active conformations of TSHR and LH/CGR reveals similar overall structures of the 7TM domain and the p10 peptide.
Extended Data Fig. 11 Cryo-EM data processing for M22-bound TSHR-Gs complex.
a) Representative image from 25,030 micrographs. Scale bar, 50 nm. b) Selected 2D class averages from final reconstruction. c) Processing approach used for reconstruction of M22-bound TSHR-Gs complex. A local resolution map was calculated from cryoSPARC using masks from indicated local refinement, then visualized with the composite map in the same scale. A viewing distribution plot was generated using scripts from the pyEM software suite and visualized in ChimeraX. GS-FSC and dFSC curves were generated in cryoSPARC and as previously described in Dang, S. et al. Nature 552, 426–j429 (2017).
Extended Data Fig. 12 Extent of ECD activation for all individual simulations across four simulation conditions.
Dashed lines indicate the projection metric values (see Methods) in the inactive (0.0 Å) and active (33.5 Å) state cryo-EM structures. Thick traces indicate the moving average smoothed over a 25-ns window, and thin traces represent unsmoothed data. Each column represents a distinct simulation condition: started from the active structure, with M22 bound (brown traces, first column); started from the active structure, with M22 removed (grey traces, second column); started from the inactive structure, with CS-17 bound (blue traces, third column); started from the inactive structure, with CS-17 removed (grey traces, fourth column).
Extended Data Fig. 13 Glycosylation of engineered K1-70glyco construct.
a) Native mass spectrum (nMS) of K1-70, K1-70glyco, and K1-70glyco(N16Q) Fab fragments. b) Accurate mass assignment of Fab fragments. nMS demonstrates ~1.7 kDa increased mass for K1-70glyco and more heterogeneity consistent with N-linked glycosylation. The smaller mass and increased homogeneity of the K1-70glyco(N16Q) construct further supports glycosylation at the engineered N16 position. c) Crystal structure of K1-70 TSHR-ECD complex (PDB: 2XWT) aligned to CS-17 bound TSHR. The membrane-proximal N229 residue (of the “GlycoB” glycosylation motif) is highlighted in red. d) cAMP production comparing K1-70 WT, K1-70 GlycoB and M22 Fab fragment-mediated activation of TSHR. Plotted data points are means of triplicate measurements ± SD from one of three biological replicates.
Supplementary information
Supplementary Table 1
Summary of TSHR signalling studies. Values are expressed as mean pEC50 or mean Emax ± s.e.m. from (n) biological replicates. ND, Not determined.
Supplementary Data 1
Structure files for static modelling of TSHR in active and inactive conformations bound to TSH, M22 CS-17, and K1-70.
Supplementary Video 1
3D variability analysis of TR1402-bound TSHR–Gs complex.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Faust, B., Billesbølle, C.B., Suomivuori, CM. et al. Autoantibody mimicry of hormone action at the thyrotropin receptor. Nature 609, 846–853 (2022). https://doi.org/10.1038/s41586-022-05159-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05159-1
This article is cited by
-
Self Fourier shell correlation: properties and application to cryo-ET
Communications Biology (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.