Abstract
Diabetes mellitus is prevalent among women of reproductive age, and many women are left undiagnosed or untreated1. Gestational diabetes has profound and enduring effects on the long-term health of the offspring2,3. However, the link between pregestational diabetes and disease risk into adulthood in the next generation has not been sufficiently investigated. Here we show that pregestational hyperglycaemia renders the offspring more vulnerable to glucose intolerance. The expression of TET3 dioxygenase, responsible for 5-methylcytosine oxidation and DNA demethylation in the zygote4, is reduced in oocytes from a mouse model of hyperglycaemia (HG mice) and humans with diabetes. Insufficient demethylation by oocyte TET3 contributes to hypermethylation at the paternal alleles of several insulin secretion genes, including the glucokinase gene (Gck), that persists from zygote to adult, promoting impaired glucose homeostasis largely owing to the defect in glucose-stimulated insulin secretion. Consistent with these findings, mouse progenies derived from the oocytes of maternal heterozygous and homozygous Tet3 deletion display glucose intolerance and epigenetic abnormalities similar to those from the oocytes of HG mice. Moreover, the expression of exogenous Tet3 mRNA in oocytes from HG mice ameliorates the maternal effect in offspring. Thus, our observations suggest an environment-sensitive window in oocyte development that confers predisposition to glucose intolerance in the next generation through TET3 insufficiency rather than through a direct perturbation of the oocyte epigenome. This finding suggests a potential benefit of pre-conception interventions in mothers to protect the health of offspring.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All the sequencing data can be viewed in NODE under accession OEP002503. All other data are available from the corresponding authors on reasonable request. Statistical details are in Supplementary Table 5. Full immunoblots are provided as Supplementary Fig. 1. Source data are provided with this paper.
References
International Diabetes Federation. IDF Diabetes Atlas, 10th edn. Brussels, Belgium: International Diabetes Federation, 2021.
Pettitt, D. J., Baird, H. R., Aleck, K. A., Bennett, P. H. & Knowler, W. C. Excessive Obesity in Offspring of Pima Indian Women with Diabetes during Pregnancy. New Engl. J. Med. 308, 242–245 (1983).
Hjort, L. et al. Diabetes in pregnancy and epigenetic mechanisms-how the first 9 months from conception might affect the child's epigenome and later risk of disease. Lancet Diabetes Endocrinol 7, 796–806 (2019).
Gu, T. P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Zimmet, P. Z., Magliano, D. J., Herman, W. H. & Shaw, J. E. Diabetes: a 21st century challenge. Lancet Diabetes & Endocrinology 2, 56–64 (2014).
Clausen, T. D. et al. High Prevalence of Type 2 Diabetes and Pre-Diabetes in Adult Offspring of Women With Gestational Diabetes Mellitus or Type 1 Diabetes The role of intrauterine hyperglycemia. Diabetes Care 31, 340–346 (2008).
Tam, W. H. et al. In Utero Exposure to Maternal Hyperglycemia Increases Childhood Cardiometabolic Risk in Offspring. Diabetes Care 40, 679 (2017).
Lowe, W. L. et al. Hyperglycemia and Adverse Pregnancy Outcome Follow-up Study (HAPO FUS): Maternal Gestational Diabetes Mellitus and Childhood Glucose Metabolism. Diabetes Care 42, 372–380 (2019).
Watson, E. D. & Rakoczy, J. Fat eggs shape offspring health. Nat. Genet. 48, 478–479 (2016).
Sales, V. M., Ferguson-Smith, A. C. & Patti, M. E. Epigenetic Mechanisms of Transmission of Metabolic Disease across Generations. Cell Metab 25, 559–571 (2017).
Harris, J. E. et al. Exercise-induced 3′-sialyllactose in breast milk is a critical mediator to improve metabolic health and cardiac function in mouse offspring. Nature Metabolism 2, 678–687 (2020).
Godfrey, K. M., Gluckman, P. D. & Hanson, M. A. Developmental origins of metabolic disease: life course and intergenerational perspectives. Trends Endocrinol. Metab. 21, 199–205 (2010).
Skinner, M. K., Manikkam, M. & Guerrero-Bosagna, C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 21, 214–222 (2010).
Rando, O. J. & Simmons, R. A. I’m eating for two: parental dietary effects on offspring metabolism. Cell 161, 93–105 (2015).
Boskovic, A. & Rando, O. J. in Annual Review of Genetics, Vol 52 Vol. 52 Annual Review of Genetics (ed N. M. Bonini) 21-41 (2018).
Ge, Z. J. et al. DNA methylation in oocytes and liver of female mice and their offspring: effects of high-fat-diet-induced obesity. Environ. Health Perspect. 122, 159–164 (2014).
Huypens, P. et al. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat. Genet. 48, 497–499 (2016).
Daxinger, L. & Whitelaw, E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13, 153–162 (2012).
Chen, Q., Yan, W. & Duan, E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 17, 733–743 (2016).
Mauvais-Jarvis, F. Estrogen and androgen receptors: regulators of fuel homeostasis and emerging targets for diabetes and obesity. Trends Endocrinol. Metab. 22, 24–33 (2011).
Muoio, D. M. & Newgard, C. B. Molecular and metabolic mechanisms of insulin resistance and β-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9, 193–205 (2008).
Kleinert, M. et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 14, 140–162 (2018).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001).
Sharma, U. & Rando, O. J. Metabolic Inputs into the Epigenome. Cell Metabolism 25, 544–558 (2017).
Cavalli, G. & Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019).
Wossidlo, M. et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241 (2011).
Iqbal, K., Jin, S.-G., Pfeifer, G. P. & Szabó, P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. 108, 3642–3647 (2011).
Tan, L. & Shi, Y. G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139, 1895 (2012).
Wang, Q., Chi, M. M., Schedl, T. & Moley, K. H. An intercellular pathway for glucose transport into mouse oocytes. Am J Physiol Endocrinol Metab 302, E1511–E1518 (2012).
Spruijt, C. G. & Vermeulen, M. DNA methylation: old dog, new tricks? Nat. Struct. Mol. Biol. 21, 949–954 (2014).
Boyes, J. & Bird, A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64, 1123–1134 (1991).
Iynedjian, P. B. Molecular physiology of mammalian glucokinase. Cell. Mol. Life Sci. 66, 27–42 (2009).
Efrat, S. et al. Ribozyme-mediated attenuation of pancreatic β-cell glucokinase expression in transgenic mice results in impaired glucose-induced insulin secretion. Proc Natl Acad Sci U S A 91, 2051–2055 (1994).
Grupe, A. et al. Transgenic knockouts reveal a critical requirement for pancreatic β cell glucokinase in maintaining glucose homeostasis. Cell 83, 69–78 (1995).
Terauchi, Y. et al. Pancreatic β-cell-specific targeted disruption of glucokinase gene. Diabetes mellitus due to defective insulin secretion to glucose. J. Biol. Chem. 270, 30253–30256 (1995).
Shen, J. & Zhu, B. Integrated analysis of the gene expression profile and DNA methylation profile of obese patients with type 2 diabetes. Molecular medicine reports 17, 7636–7644 (2018).
Joglekar, M. V. et al. Expression of miR-206 in human islets and its role in glucokinase regulation. American journal of physiology. Endocrinology and metabolism 315, E634–e637 (2018).
Cauchi, S. et al. European genetic variants associated with type 2 diabetes in North African Arabs. Diabetes & metabolism 38, 316–323 (2012).
Bonnefond, A. et al. Pathogenic variants in actionable MODY genes are associated with type 2 diabetes. Nature Metabolism 2, 1126–1134 (2020).
Terauchi, Y. et al. Glucokinase and IRS-2 are required for compensatory β cell hyperplasia in response to high-fat diet-induced insulin resistance. J. Clin. Invest. 117, 246–257 (2007).
Lu, B. et al. Impaired β-cell glucokinase as an underlying mechanism in diet-induced diabetes. Dis. Models Mech. 11, dmm033316 (2018).
Tang, L. et al. Elevated CpG island methylation of GCK gene predicts the risk of type 2 diabetes in Chinese males. Gene 547, 329–333 (2014).
Dhliwayo, N., Sarras, M. P. Jr, Luczkowski, E., Mason, S. M. & Intine, R. V. Parp inhibition prevents ten-eleven translocase enzyme activation and hyperglycemia-induced DNA demethylation. Diabetes 63, 3069–3076 (2014).
Wu, D. et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 559, 637–641 (2018).
Zhang, Q. et al. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J. Biol. Chem. 289, 5986–5996 (2014).
Yuan, E.-F. et al. Hyperglycemia affects global 5-methylcytosine and 5-hydroxymethylcytosine in blood genomic DNA through upregulation of SIRT6 and TETs. Clinical Epigenetics 11, 63 (2019).
Pinzon-Cortes, J. A. et al. Effect of diabetes status and hyperglycemia on global DNA methylation and hydroxymethylation. Endocr Connect 6, 708–725 (2017).
Deeds, M. C. et al. Single dose streptozotocin-induced diabetes: considerations for study design in islet transplantation models. Lab. Anim. 45, 131–140 (2011).
Krishnamurthy, J. et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443, 453–457 (2006).
Tesch, G. H. & Allen, T. J. Rodent models of streptozotocin-induced diabetic nephropathy (Methods in Renal Research). Nephrol. 12, 261–266 (2007).
Huan, L. et al. GDF11 Attenuates development of type 2 diabetes via improvement of islet β-cell function and survival. Diabetes 66, 1914–1927 (2017).
Bohacek, J. & Mansuy, I. M. A guide to designing germline-dependent epigenetic inheritance experiments in mammals. Nat. Methods 14, 243–249 (2017).
Behringer, R., Gertsenstein, M., Nagy, K. V. & Nagy, A. (eds) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2003).
Ayala, J. E. et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model. Mech. 3, 525–534 (2019).
Zhu, H. et al. Insulin therapy for gestational diabetes mellitus does not fully protect offspring from diet-induced metabolic disorders. Diabetes 68, 696–708 (2019).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Harris, S. E., Gopichandran, N., Picton, H. M., Leese, H. J. & Orsi, N. M. Nutrient concentrations in murine follicular fluid and the female reproductive tract. Theriogenology 64, 992–1006 (2005).
Gu, C., Liu, S., Wu, Q., Zhang, L. & Guo, F. Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res 29, 110–123 (2019).
Smallwood, S. A. et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014).
Guo, W. et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14, 774 (2013).
Guo, W. et al. CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics 34, 381–387 (2018).
Krueger, F. & Andrews, S. R. SNPsplit: Allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Res 5, 1479 (2016).
Schutsky, E. K. et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat Biotechnol (2018).
Guo, F. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 (2014).
Shen, L. et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–471 (2014).
Adenot, P. G., Mercier, Y., Renard, J. P. & Thompson, E. M. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development (Cambridge, England) 124, 4615–4625 (1997).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Yang, H., Wang, H. Y. & Jaenisch, R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9, 1956–1968 (2014).
Dai, H. Q. et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 538, 528-+ (2016).
Coope, G. J. et al. Predictive blood glucose lowering efficacy by Glucokinase activators in high fat fed female Zucker rats. Br. J. Pharmacol. 149, 328–335 (2006).
Grimsby, J. et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science 301, 370–373 (2003).
Wang, P. et al. Effects of a Novel Glucokinase Activator, HMS5552, on Glucose Metabolism in a Rat Model of Type 2 Diabetes Mellitus. J Diabetes Res 2017, 5812607 (2017).
Gorman, T. et al. Effect of high-fat diet on glucose homeostasis and gene expression in glucokinase knockout mice. Diabetes Obes Metab 10, 885–897 (2008).
Johnson, D. et al. Glucose-dependent modulation of insulin secretion and intracellular calcium ions by GKA50, a glucokinase activator. Diabetes 56, 1694–1702 (2007).
Wei, P. et al. Effects of glucokinase activators GKA50 and LY2121260 on proliferation and apoptosis in pancreatic INS-1 β cells. Diabetologia 52, 2142–2150 (2009).
Acknowledgements
We thank B. Han for providing APOBEC3A deaminase, Z. Shen for anti-GCK antibody; S. Hong, C. Zhou and G. Xu for technical assistance in embryo manipulation; M. Cai for data analysis; L. Shen, Y. Pan, H. Wang, Q. Xu, L. Xu, J. Xue, Z. Zhou, X. Liu, C. Xu, J. Zhang and H. Yang for discussions; and T. Gu, L. Rui, X. Li, Y. Chen, H. Fan and S. Xuan for critical reading of the manuscript. This work is supported by the National Key R&D Program of China (2017YFC1001300 to H.H.; 2018YFA0800302 to G.-L.X.; 2017YFC1001301 to Y.-R.D.; and 2018YFC1004402 to J.S.), the National Science Foundation of China (82088102 to H.H.; 81661128010 to H.H.; 31991163 to G.-L.X.; 31671569 to J.S.; and 81971458 to G.D.), Shanghai Jiao Tong University, CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-064 to H.H.), Collaborative Innovation Program of Shanghai Municipal Health Commission (2020CXJQ01 to H.H.), Technology Innovation Action Plan Hong Kong, Macao and Taiwan Science and Technology Cooperation Project (19410760100 to H.H.), Shanghai Frontiers Science Research Base of Reproduction and Development (to H.H.) and Zhejiang Provincial Natural Science Foundation of China under Grant (LQ21H040005 to B.C.).
Author information
Authors and Affiliations
Contributions
H.H. conceived the original idea and, together with G.-L.X. J.S., B.C. and Y.-R.D. designed the experiments. Experiments were performed by B.C. with the help of: Y.-R.D. for the experiments associated with oocytes and RT–qPCR; H.Z. for mouse modelling, phenotyping and human sample collection; M.-L.S. for the experiments associated with TET3 rescue, allele-specific methylation measurement and mouse modelling; C.W. for Smart-seq2 and PBAT library building, data analysis and bisulfite sequencing; Y.C. for islet phenotyping and mouse modeling; H.P. for pyrosequencing analysis; G.D. for supervising human sample collection; J.G. for micromanipulation; Y.T. for human oocyte IVM; and Z.-M.X. for animal housing and genotyping. Luyang Jin, Y. Zhou and C.Y. assisted with mouse modelling and phenotyping. P.R.F. assisted with interpretation of data. X.T., P.L., F.Z., Q.Z., L.W., D.L., Y.Y., Li Jin, S.Z., Y. Zhu, X.L. and Y.W. recruited the patients and collected the human samples. B.C., Y.-R.D., P.R.F., G.-L.X. and H.H. wrote the manuscript with contributions from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Evan Rosen and the other, anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 STZ-induced hyperglycemia leads to reduced oocyte retrieval but does not affect the development potential of early embryos resulted.
a, Gradually increased blood glucose levels after STZ treatment in female mice. Ctrl, HG: n = 16 and 26 mice, respectively. MII oocytes were collected for studies 30 days upon STZ injection (HG oocytes). b–c, Reduced GV (b) and MII (c) oocytes retrieval per mouse. Ctrl, HG: n = 6 mice per group for GV, and n = 14 independent experiments per group for MII, respectively. d, Granulosa cell dysfunction in HG mice evidenced by decreased serum estradiol (left) but increased free testosterone (right) concentration 48 h after PMSG priming. Ctrl, HG: n = 8 and 6 mice for estradiol; n = 8 and 8 mice for free testosterone, respectively. e, Smaller cumulus-oocyte complex (COC) in HG mice. Shown are representative images of COCs retrieved from control and hyperglycemic mice in 3 independent experiments with similar results. Scale bar, 500 μm. f, HG zygotes did not exhibit developmental delay at the timepoint 6 h post IVF when PN3–4 male pronuclei were collected. g–h, Maternal hyperglycemia did not significantly affect the 2-cell embryo formation (g) or blastocyst formation (h) in the IVF experiments. Ctrl, HG: n = 13 independent experiments per group. i, Representative images of oocytes and IVF-derived pre-implantation embryos in HG versus control group in 3 independent experiments with similar results. Scale bar, 100 μm. j, Normal litter size of offspring born from transferred 2-cell embryos derived from HG oocytes. Ctrl, HG: n = 20 and 23 deliveries, respectively. k, Normal appearance of HG E18.5 fetuses and placentas. Representative litters are shown. Scale bar, 2 cm. Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by the unpaired (b, d, j) or paired (c, g, h) Student’s t-test. *P < 0.05, *P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
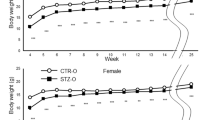
Extended Data Fig. 2 Age-dependent and HFD-accelerated progression of glucose intolerance in both sexes of HG offspring.
a, Normal body weight trajectories of HG offspring. Males and females were separately calculated from weaning at 3 weeks of age. Ctrl, HG: n = 18 and 14 mice for males; 12 and 17 mice for females, respectively. b, GTT results of female offspring at 16 weeks of age. c–f, GTT results of male (c) and female (e) offspring at the indicated ages and the corresponding area under the curves (AUCs) of glucose levels (d, f). GTT results of 16-week-old offspring are shown in Fig. 1b and Extended Data Fig. 2b. Experiments were independently repeated on 2 offspring cohorts with similar results. 8 weeks to 1 year: Ctrl, HG: n = 16 and 12 mice for males; 13 and 12 mice for females, respectively. 2 years: Ctrl, HG: n = 13 and 11 mice for males; n = 12 and 11 mice for females, respectively. g, Normal body weight trajectories of HG offspring on HFD from 3 weeks on. Males and females were separately calculated from weaning at 3 weeks of age and were fed a HFD from then on. Ctrl, HG: n = 11 and 13 mice for males; n = 11 and 12 mice for females, respectively. h–i, Glucose intolerance in both sexes after HFD challenge. Shown are GTT results of male (h) and female (i) offspring at 12 weeks of age and the corresponding AUCs (j). Ctrl, HG: n = 11 and 13 mice for males; n = 11 and 12 mice for females, respectively. a–j, Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test (b, c, e, h–i) or the unpaired Student’s t-test (a, d, f, g, j). *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 3 Glucose intolerance phenotype does not persist to F2 generation.
a, Schematic diagram of the breeding for F2 offspring. Circles designate females and squares designate males. F2 were generated by intercrossing F1 males and females from control and pregestational hyperglycemic mothers. b, GTT results of male (left) and female (right) offspring at 24 weeks of age. c, Area under the curve (AUC) of the blood glucose levels in male and female. b–c, Ctrl♂×Ctrl♀, Ctrl♂×HG♀, HG♂×Ctrl♀, HG♂×HG♀: n = 8 mice per group for both males and females. Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by one-way repeated-measurements ANOVA (b) or one-way ANOVA (c). Statistical details are in Supplementary Table 5.
Extended Data Fig. 4 Normal insulin sensitivity and pancreas histomorphometry but defective insulin secretion in HG offspring.
a–d, Normal insulin sensitivity in HG offspring fed either chow diet or HFD. Shown are ITT results of male (a) and female (b) with a chow diet, and male (c) and female (d) offspring after HFD induction at 16 weeks of age. a–d, Ctrl, HG: n = 16 and 12 mice for males; 13 and 12 mice for females. Ctrl-HFD and HG-HFD: n = 9 and 13 mice for males; 7 and 12 mice for females, respectively. e, The stimulation index of insulin release from isolated pancreatic islets of female offspring after HFD feeding at 24 weeks of age. Ctrl, HG: n = 9 per mouse. f–h, Insulin immunohistochemistry (IHC) (f), Hematoxylin and eosin (H&E) staining (g) and immunofluorescence (IF) (h) in paraffin-embedded pancreas sections from 16-week-old male offspring. Shown are representative images from 5 mice per group with similar results. No significant differences of islets in size and distribution (f), morphology (g) or α/β cell ratio (h) were detected between control and HG. Scale bar, 1 mm (f), 50 μm (g–h). i–k, Analysis of pancreatic morphology from 16-week-old male offspring by H&E staining. Ctrl, HG: n = 11 and 9 mice, respectively (with 2 sections per mouse). i, Average islet size. j, Islet density expressed as the average number of islets per 107 μm2 of pancreas area. k, Profiling of islet size distribution. l, Total pancreatic insulin content of 16-week-old male offspring. Ctrl, HG: n = 9 mice per group. a–e, i–l, Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test (a–d) and the unpaired Student’s t-test (e, i–l). **P < 0.01. Statistical details are in Supplementary Table 5.
Extended Data Fig. 5 Reduced Tet3 expression in mouse oocytes and zygotes, and impaired 5mC oxidation in the zygotic DNA of one-cell embryos from HG females.
a, Principal component analysis (PCA) of the transcriptional profile of MII oocytes identifies a clear separation between control and HG group. b, Scatter plot shows the differentially expressed genes (DEGs) of HG MII oocytes compared to controls. nCount stands for normalized counts (mean of the counts divided by size factors; DESeq2 software package). The dashed lines indicate the 1.5-fold change threshold for identifying DEGs. Red and blue dots denote significantly changed genes (padj < 0.05 and fold change ≥ 1.5) and grey dots denote not significantly changed genes. c, Gene ontology analysis of the downregulated genes in HG MII oocytes from RNA-seq. The red vertical line corresponds to P = 0.05. d, Expression of DNA-modifying enzymes in MII oocytes determined by RNA-seq. Tet3 expression is shown in Fig. 2b. e, Validation of the relative gene expression levels in MII oocytes from HG females by RT-qPCR. Gene expression shown in the left panel is the qPCR result of abundantly expressed genes encoding DNA-modifying enzymes in 10 pooled oocytes using the method of Smart-seq2 pre-amplification. Ctrl and HG: n = 8 and 9 replicates, respectively. The right panel is the confirmation of relative Tet3 expression in 60 pooled oocytes independently isolated using direct RT-qPCR relative to 18S rRNA. f, The HFD-fed and low-dose STZ‑induced type 2 diabetes mellitus (T2DM) mice of 16-week-old showed obesity and increased blood glucose when oocytes were collected. Ctrl, T2DM: n = 14 and 9 mice, respectively. g, Decreased Tet3 mRNA expression in GV oocytes from 16-week-old T2DM mice versus control by RT-qPCR. Ctrl, T2DM: n = 14 and 9 mice (with 5 oocytes pooled per mouse), respectively. h–k, Representative images of anti-TET3 (magenta) and DAPI (blue) staining in mouse zygotes at PN3–4 stage (h) and the quantification of relative fluorescence intensity levels (j). Ctrl, HG: n = 8 and 10, respectively. Representative images of anti-5hmC (red) and anti-5mC (green) staining in mouse zygotes at PN3–4 stage (i) and the quantification of relative fluorescence intensity levels of 5hmC and 5mC (k). Ctrl, HG: n = 15 and 16, respectively. Male and female pronuclei are indicated by their respective sex symbols. PB, polar body. PN, pronucleus. Scale bars, 10 μm. Each data point is based on the level of the TET3 signal relative to the DAPI staining intensity (j) or 5hmC relative to 5mC (k) in the same pronucleus. b–c, P values were calculated by Wald test corrected for multiple testing using the Benjamini and Hochberg method by default (b) and Fisher's Exact test (c). d–g, j–k, Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by the unpaired Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 6 Elevated glucose concentration in follicles and decreased Tet3 mRNA levels in GV and MII oocytes in response to high-glucose concentrations.
a, Classification criteria of mice that underwent STZ injection according to the nonfasting blood glucose level on the day of IVF. b, Nonfasting blood glucose range of 8-week-old female rats during the day of follicle collection (left) and high follicular fluid glucose concentrations measured in HG rats (right). n = 3 replicates per group (with fluid from antral follicles collected from 3 rats pooled for each replicate). c, The upper schematic diagram shows the IVG process of primary follicles for 12 days until the oocytes reach the fully-grown GV stage with or without high-glucose treatment. The lower panel shows Tet3 mRNA expression in GV oocytes from IVG under the indicated glucose conditions. For the 5.5 mM, 30 mM D-glucose, 5.5 mM D-glucose + 24.5 mM L-glucose: n = 14, 17 and 14 oocytes, respectively. d, The upper schematic diagram shows the IVM process for 16 h from GV oocytes to MII oocytes with or without high-glucose treatment. The lower panel shows Tet3 mRNA expression in mouse MII oocytes from IVM under the indicated glucose conditions. For the 5 mM, 15 mM, 30 mM D-glucose, 5 mM D-glucose + 25 mM L-glucose groups: n = 10, 9, 10 and 8 replicates, respectively (with 10 oocytes pooled for each replicate). e, The upper schematic diagram of mRNA micro-injection and oocyte maturation to investigate whether high glucose condition leads to the degradation of Tet3 transcript through its 3’-UTR. GFP transcript fused with the Tet3 3’-UTR, mCherry with Tubb3 3’-UTR (an unaffected gene as an internal control), BFP with polyA (normalization control) mRNA were co-injected into GV oocytes. The lower panel shows the relative mRNA levels determined by RT-qPCR. Ctrl (5.5 mM), 30 mM: n = 16 per group. f, Representative images of MII oocytes after IVM at various glucose levels from 2 independent experiments with similar results. Scale bar, 200 μm. g, Human follicular fluid glucose concentrations measured in the patients with or without diabetes. Diabetic patients were recruited according to the current diagnosis of diabetes (according to ref. 1). Control and Diabetes: n = 14 humans per group. h, TET3 mRNA expression in human MII oocytes from IVM under the indicated glucose conditions. For the 2.5 mM, 10 mM and 15 mM groups, n = 9, 9 and 11 oocytes, respectively. b–h, Data are shown as scatter dot plots (b, e, g, mean ± s.e.m.), and box-and-whisker plots (c, d, h, the median (middle lines), interquartile range (boxes) and minimum to maximum values (whiskers)). Two-tailed P values were calculated by the one-way ANOVA with the post hoc LSD multiple comparisons (c, d, h) or the unpaired Student’s t-test (b, e, g). *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 7 Analysis of methylation alterations in HG E18.5 fetal islets by whole-genome bisulfite sequencing.
a, Methylation levels of Ctrl and HG E18.5 fetal islets across different genomic elements. Methylation level was calculated using CpG sites with at least 3× sequencing coverage. CGI, CpG island. b, Methylome comparison between Ctrl and HG islets using DMRs. DMRs were filtered by length (≤ 1000 bp) and CpG number (at least 5 CpG sites). Maximal distance betweentwo adjacent cytosines in CpG was 100 bp. Those with at least 20% absolute methylation level difference between HG and Ctrl samples were defined as hypermethylated or hypomethylated DMRs. Each dot represents a single DMR. c, Distribution of hyper-DMRs present in HG islets across various genomic features. For regions belonging to more than one category, the classification followed the priority of promoter, exon, intron and intergenic regions. d, A list of genes from the insulin secretion pathway with hyper-DMRs present in their promoter region. * denotes that these DMRs are hypermethylated only in the paternal allele (4/9). † denotes that these DMRs are hypermethylated in both alleles (1/9). ‡ denotes that the parental origin of these hyper-DMRs is indistinguishable (4/9). The numbers inside parentheses denote the count of genes belong to each category relative to the total gene count. P values were calculated by Fisher’s exact test.
Extended Data Fig. 8 Epigenetic inheritance of hypermethylation at the Gck promoter throughout HG offspring development.
a, BS analysis of H19 imprinted control region methylation in sperm, oocytes, male and female pronuclei isolated from aphidicolin-treated PN3–4 zygotes. Fully methylated H19 in sperm and male pronuclei as well as unmethylated H19 in oocytes and female pronuclei indicate high purity of germ cells and pronuclei without somatic cell contamination. b, BS analysis of Line 1 5’ region methylation in the sperm and male pronuclei. Hypermethylation at 5’ Line 1 elements suggests compromised Tet3-mediated demethylation in the HG group. c–d, BS analysis of the Gck promoter methylation in sperm, MII oocytes (c) and female pronuclei (d) of the indicated groups. Relatively high levels of methylation in sperm and low levels of methylation in MII oocytes and female pronuclei at the Gck promoter were observed. e, Schematic diagram of the strategy to distinguish Gck methylation patterns on the paternal allele from maternal allele. sgRNA targeting sequence and PAM sequence are underlined and labeled in red, respectively. Through CRISPR/Cas9 mediated editing, a homozygous mutant male with a 35 bp-deletion before the Gck promoter was generated. PCR primers for bisulfite methylation analysis are also indicated. The forward primers used are allele-specific. f, Bisulfite sequencing analysis of promoter methylation of the paternal (left) and maternal (right) Gck in aphidicolin-treated PN5 zygotes using the strategy described in e. a–f, Open and filled circles represent unmethylated and methylated CpG sites, respectively. g–j, Pyrosequencing analysis of the Gck promoter methylation in pancreatic islets from offspring at 3 weeks (male: g, female: h) and 16 weeks (male: i, female: j) of age. n of the groups is indicated. k, Timeline of the average methylation level of the Gck corresponding sites in Figure 3c–e and Extended Data Fig. 8g–j. Only the percentages of 5mC were included in the methylation level in male pronuclei. l, Reduced Gck mRNA expression in female offspring pancreatic islets. n = 9 mice per group. m, Relative mRNA expression of genes with hyper-DMRs involved in insulin secretion. RT-qPCR analysis was performed for pancreatic islets from 3-week-old male offspring. Ctrl, HG: n = 11 and 12 mice, respectively. n–p, Using glucokinase activator (GKA) Dorzagliatin to reverse the effect of Gck downregulation in HG offspring. n, Glucose tolerance test (GTT) results of male offspring at 24 weeks of age after HFD feeding. Ctrl, HG, and HG + GKA: n = 8 mice per group. P values for comparisons of HG + GKA and HG are presented using *, HG and Ctrl using †. o, The stimulation index of insulin release from isolated pancreatic islets of male offspring at 24 weeks of age. Ctrl, HG and HG + GKA: n = 6, 9 and 7 mice, respectively. p, Total insulin released based on the corresponding data in o. g–p, Data are presented as the bar charts and line charts (g–l, n–p, mean ± s.e.m.) and box-and-whisker plots (m, the median (middle lines), interquartile range (boxes) and minimum to maximum values (whiskers)); two-tailed P values were calculated by one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test (n) or the unpaired Student’s t-test (g–m, o–p). *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 9 Analysis of allelic origin of the methylation alterations in HG offspring.
a–b, Pyrosequencing analysis of paternal (a) and maternal (b) Gck promoter methylation in blastocysts using the strategy described in Extended Data Fig. 8e. Ctrl, HG: n = 8 and 7 replicates (with 5 blastocysts pooled for each sample). c, Average methylation levels in a and b. d, Allelic methylation profile of promoter hyper-DMRs in the 4 indicated genes of the insulin secretion pathway based on WGBS data. Pat, paternal. Mat, maternal. Vertical bars (overall, grey; paternal, blue; maternal, pink) above the horizontal line indicate the methylation level (0–1) at individual CpG dyads (counting the two complementary CpGs), and light grey bars below the horizontal line denote the CpGs detected as unmethylated to distinguish them from undetected CpG sites. Dash line boxes indicate hyper DMRs for analysis of their parental origin. a–c, Data are presented as mean ± s.e.m.; two-tailed P values were calculated by the unpaired Student’s t-test (a–c). *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 10 Increased methylation levels at the Line 1 5’ region and the Gck promoter and impaired glucose tolerance in the offspring from oocyte-specific Tet3-conditional gene knockout dams.
a–b, Illustrative diagram of experimental groups with reduced levels of maternal TET3 expression induced by maternal hyperglycemia (a) and oocyte-specific Tet3 heterozygous and homozygous knockout (b). c, BS analysis of the H19 imprinted control region in the sperm and male pronuclei isolated from aphidicolin-treated zygotes of PN3–4 stage. d, BS analysis of the Line 1 5’ region in the sperm and male pronuclei from aphidicolin-treated PN3–4 zygotes. e, BS analysis of the Gck promoter region in the female pronuclei isolated from aphidicolin-treated zygotes of PN3–4 stage. c–e, Open and filled circles represent unmethylated and methylated CpG sites, respectively. The percentages of methylated CpGs are indicated. f–g, Pyrosequencing analysis of the Gck promoter in pancreatic islets from offspring at 3 weeks of age (male: f, female: g). n of the group is indicated. P values for comparisons of Mat-Het and Mat-WT are presented using *, Mat-KO and Mat-WT using †. h, Gck mRNA expression in female pancreatic islets at 3 weeks of age. Mat-WT, Mat-Het, Mat-KO: n = 8, 10, 11 mice, respectively. i, Western blot analysis of GCK in pancreatic islets of 3-week-old offspring males. Each sample was pooled islets from 3–5 mice. Gel source data are presented in Supplementary Figure 1. j–k, GTT results of female offspring at 16 weeks of age (j) and the corresponding AUCs (k). Mat-WT, Mat-Het, Mat-KO: n = 11, 13 and 12 mice, respectively. P values for comparisons of Mat-Het and Mat-WT are presented using *, Mat-KO and Mat-WT using †. f–k, Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test with Mat-WT (j) and the unpaired Student’s t-test with Mat-WT (f–h, k). *P < 0.05, **P < 0.01 and ***P < 0.001; ††P < 0.01 and †††P < 0.001. Statistical details are in Supplementary Table 5.
Extended Data Fig. 11 Exogeneous TET3 expression in HG MII oocytes suppresses the maternal effect on DNA methylation and offspring susceptibility to metabolic impairments.
a, Schematic diagram of TET3 rescue experiment relying on mRNA injection into HG MII oocytes. b, Representative images of 5mC (green) and 5hmC (red) staining in Mat-WT, Mat-KO and HG zygotes at PN5 stage which were derived from oocytes that were injected with H2O, wildtype Tet3 or Tet3-mut mRNA at MII stage. Male and female pronuclei are indicated by their respective sex symbols. Scale bar, 10 μm. c, BS analysis of the H19 imprinted control region in the male and female pronuclei isolated from aphidicolin-treated zygotes of PN3–4 stage. d, BS analysis of the Gck promoter region in the female pronuclei isolated from aphidicolin-treated zygotes of PN3–4 stage. c–d, Open and filled circles represent unmethylated and methylated CpG sites, respectively. e–f, Pyrosequencing analysis of the Gck promoter methylation in mouse blastocysts (e) and average methylation levels in e (f). Mat-KO + Tet3-mut, Mat-KO + Tet3: n = 6 and 7 replicates (with 5 blastocysts pooled for each replicate). g, Average methylation levels in Figure 4g. Ctrl, HG + Tet3, HG + Tet3-mut: 4, 5 and 4 replicates (with 5 blastocysts pooled for each replicate), respectively. h, Gck mRNA expression in pancreatic islets of male offspring at 1 year of age. Ctrl, HG + Tet3, HG + Tet3-mut: n = 8, 8 and 10 mice, respectively. i, Western blot analysis of GCK in pancreatic islets of offspring males at 1 year of age. Each sample was pooled islets from 2–3 mice. Gel source data are presented in Supplementary Figure 1. j–k, GTT of male offspring at 24 weeks of age (j) and the corresponding AUCs (k). Mat-KO + Tet3-mut, Mat-KO + Tet3: n = 12 and 14 mice. l, GTT of male offspring at 24 weeks of age after HFD feeding. Ctrl, HG + Tet3, HG + Tet3-mut: n = 10, 12 and 11 mice, respectively. P values for comparisons of HG + Tet3-mut and Ctrl are presented using *, HG + Tet3 and HG + Tet3-mut using †. m, The corresponding AUCs of the blood glucose level in Figure 4h and Extended Data Fig. 11l. e–m, Data are presented as the mean ± s.e.m.; two-tailed P values were calculated by the unpaired Student’s t-test (e–f, k) or one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test (k), one-way repeated-measurements ANOVA followed by the post hoc unpaired t-test with HG + Tet3-mut (l) and one-way ANOVA followed by the unpaired Student’s t-test with HG + Tet3-mut (g, h, m). *P < 0.05, **P < 0.01 and ***P < 0.001; †P < 0.05. Statistical details are in Supplementary Table 5.
Supplementary information
Supplementary Figure 1
Uncropped images of western blots. This file contains full scanned uncropped images for western blots (related to Extended Data Figs.10i, 11i), with the molecular weight markers indicated. Red boxes show the cropped bands.
Supplementary Table 1
Expression profiles of MII oocytes from control and hyperglycaemic mice by RNA-seq. This file contains normalized raw expression and statistical analysis of gene transcription for control and HG MII oocytes.
Supplementary Table 2
Characteristics of human subjects. This file contains the detailed information of the involved patients related to Figs. 2d, 3g and Extended Data Fig. 6h.
Supplementary Table 3
Analysis of promoter-associated DMRs based on the methylome generated by WGBS. This file contains a list of DMRs at gene promoters in E18.5 HG fetal islets identified by WGBS, with their gene coordinate, location within the genome, the distance to the nearest gene and statistical analysis.
Supplementary Table 4
Primer sequences. This file shows the primer sequences for RT–qPCR, bisulfite and ACE Sanger sequencing, pyrosequencing and the introduction of a 35-bp deletion before the Gck promoter by CRISPR–Cas9.
Supplementary Table 5
Detailed statistical information. This file shows the statistical methods used and exact P-values shown in the figures.
Source data
Rights and permissions
About this article
Cite this article
Chen, B., Du, YR., Zhu, H. et al. Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature 605, 761–766 (2022). https://doi.org/10.1038/s41586-022-04756-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04756-4
This article is cited by
-
Appropriate glycemic management protects the germline but not the uterine environment in hyperglycemia
EMBO Reports (2024)
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular Biology (2024)
-
Maternal Western diet mediates susceptibility of offspring to Crohn’s-like colitis by deoxycholate generation
Microbiome (2023)
-
High-fat diet in early life triggers both reversible and persistent epigenetic changes in the medaka fish (Oryzias latipes)
BMC Genomics (2023)
-
Adiposity and lipid metabolism indicators mediate the adverse effect of glucose metabolism indicators on oogenesis and embryogenesis in PCOS women undergoing IVF/ICSI cycles
European Journal of Medical Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.