Abstract
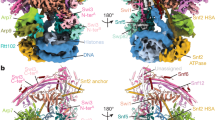
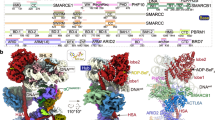
DNA wraps around the histone octamer to form nucleosomes1, the repeating unit of chromatin, which create barriers for accessing genetic information. Snf2-like chromatin remodellers couple the energy of ATP binding and hydrolysis to reposition and recompose the nucleosome, and have vital roles in various chromatin-based transactions2,3. Here we report the cryo-electron microscopy structure of the 12-subunit human chromatin-remodelling polybromo-associated BRG1-associated factor (PBAF) complex bound to the nucleosome. The motor subunit SMARCA4 engages the nucleosome in the active conformation, which reveals clustering of multiple disease-associated mutations at the interfaces that are essential for chromatin-remodelling activity. SMARCA4 recognizes the H2A–H2B acidic pocket of the nucleosome through three arginine anchors of the Snf2 ATP coupling (SnAc) domain. PBAF shows notable functional modularity, and most of the auxiliary subunits are interwoven into three lobe-like submodules for nucleosome recognition. The PBAF-specific auxiliary subunit ARID2 acts as the structural core for assembly of the DNA-binding lobe, whereas PBRM1, PHF10 and BRD7 are collectively incorporated into the lobe for histone tail binding. Together, our findings provide mechanistic insights into nucleosome recognition by PBAF and a structural basis for understanding SMARCA4-related human diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Coordinates and Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank and Protein Data Bank under accession codes EMD-31926, 7VDV (PBAF–NCP complex); EMD-31925, 7VDT (the motor–NCP complex); EMD-31927 (NBL–DBL); and EMD-31928 (NBL–HBL). Source data are provided with this paper.
References
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389, 251–260 (1997).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Yan, L. & Chen, Z. A unifying mechanism of DNA translocation underlying chromatin remodeling. Trends Biochem. Sci. 45, 217–227 (2020).
Alfert, A., Moreno, N. & Kerl, K. The BAF complex in development and disease. Epigenetics Chromatin 12, 19 (2019).
Kwon, H., Imbalzano, A. N., Khavari, P. A., Kingston, R. E. & Green, M. R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370, 477–481 (1994).
Wang, W. et al. Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J. 15, 5370–5382 (1996).
Sokpor, G., Xie, Y., Rosenbusch, J. & Tuoc, T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 10, 243 (2017).
Hodges, C., Kirkland, J. G. & Crabtree, G. R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect Med. 6, a026930 (2016).
Mittal, P. & Roberts, C. W. M. The SWI/SNF complex in cancer—biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448 (2020).
Sundaramoorthy, R. & Owen-Hughes, T. Chromatin remodelling comes into focus. F1000Res 9, https://doi.org/10.12688/f1000research.21933.1 (2020).
Pan, J. et al. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat. Genet. 51, 618–626 (2019).
Nakayama, R. T. et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet. 49, 1613–1623 (2017).
Lemon, B., Inouye, C., King, D. S. & Tjian, R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414, 924–928 (2001).
Miao, D. et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018).
Pan, D. et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018).
Ho, P. J., Lloyd, S. M. & Bao, X. Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development. Development 146, dev178780 (2019).
Leschziner, A. E., Lemon, B., Tjian, R. & Nogales, E. Structural studies of the human PBAF chromatin-remodeling complex. Structure 13, 267–275 (2005).
Mashtalir, N. et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817.e824 (2020).
He, S. et al. Structure of nucleosome-bound human BAF complex. Science 367, 875–881 (2020).
Mashtalir, N. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288.e1220 (2018).
Patsialou, A., Wilsker, D. & Moran, E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80 (2005).
Charlop-Powers, Z., Zeng, L., Zhang, Q. & Zhou, M. M. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 20, 529–538 (2010).
Sun, H. et al. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem. Biophys. Res. Commun. 358, 435–441 (2007).
Valencia, A. M. et al. Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 179, 1342–1356.e1323 (2019).
Ye, Y. et al. Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019).
He, Z., Chen, K., Ye, Y. & Chen, Z. Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity. Cell Discov. 7, 28 (2021).
Wagner, F. R. et al. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579, 448–451 (2020).
Han, Y., Reyes, A. A., Malik, S. & He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 579, 452–455 (2020).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Yan, L., Wang, L., Tian, Y., Xia, X. & Chen, Z. Structure and regulation of the chromatin remodeller ISWI. Nature 540, 466–469 (2016).
Liu, X., Li, M., Xia, X., Li, X. & Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 (2017).
Yan, L., Wu, H., Li, X., Gao, N. & Chen, Z. Structures of the ISWI–nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 26, 258–266 (2019).
Li, M. et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 567, 409–413 (2019).
Hodges, H. C. et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat. Struct. Mol. Biol. 25, 61–72 (2018).
Dykhuizen, E. C. et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497, 624–627 (2013).
Sen, P., Ghosh, S., Pugh, B. F. & Bartholomew, B. A new, highly conserved domain in Swi2/Snf2 is required for SWI/SNF remodeling. Nucleic Acids Res. 39, 9155–9166 (2011).
Sen, P. et al. The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol. Cell. Biol. 33, 360–370 (2013).
Baker, R. W. et al. Structural insights into assembly and function of the RSC chromatin remodeling complex. Nat. Struct. Mol. Biol. 28, 71–80 (2021).
McGinty, R. K. & Tan, S. Principles of nucleosome recognition by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 71, 16–26 (2021).
Wang, L., Chen, K. & Chen, Z. Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome. Nat. Commun. 12, 4057 (2021).
Gamarra, N., Johnson, S. L., Trnka, M. J., Burlingame, A. L. & Narlikar, G. J. The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h. eLife 7, e35322 (2018).
Bacic, L. et al. Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome. eLife 10, e71420 (2021).
Dao, H. T., Dul, B. E., Dann, G. P., Liszczak, G. P. & Muir, T. W. A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol. 16, 134–142 (2020).
Emery, P., Durand, B., Mach, B. & Reith, W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 24, 803–807 (1996).
Jiang, H. et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via DNMT1–Snail axis. Proc. Natl Acad. Sci. USA 117, 4770–4780 (2020).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Gao, W., Li, W., Xiao, T., Liu, X. S. & Kaelin, W. G. Jr Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc. Natl Acad. Sci. USA 114, 1027–1032 (2017).
Allen, M. D., Freund, S. M., Zinzalla, G. & Bycroft, M. The SWI/SNF subunit INI1 contains an N-terminal winged helix DNA binding domain that is a target for mutations in schwannomatosis. Structure 23, 1344–1349 (2015).
Chabanon, R. M., Morel, D. & Postel-Vinay, S. Exploiting epigenetic vulnerabilities in solid tumors: novel therapeutic opportunities in the treatment of SWI/SNF-defective cancers. Semin. Cancer Biol. 61, 180–198 (2020).
Barisic, D., Stadler, M. B., Iurlaro, M. & Schubeler, D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 569, 136–140 (2019).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. https://doi.org/10.1385/1-59259-681-9:1 (1999).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Chen, Z. L. et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Wang, N. et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell 184, 370–383.e313 (2021).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Patel, A. B. et al. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. eLife 8, e54449 (2019).
Bastiray, A., Giri, M. & Singh, M. Sequential backbone resonance assignment of AT-rich interaction domain of human BAF200. Biomol. NMR Assign. 13, 115–119 (2019).
Du, Z. et al. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 16, 5634–5651 (2021).
Zhou, N., Wang, H. & Wang, J. EMBuilder: a template matching-based automatic model-building program for high-resolution cryo-electron microscopy maps. Sci Rep. 7, 2664 (2017).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Makde, R. D., England, J. R., Yennawar, H. P. & Tan, S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010).
Acknowledgements
We thank the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for the cryo-EM facility. This work was supported by the National Key Research and Development Program (2019YFA0508902 to Z.C.), the National Natural Science Foundation of China (32130016 and 31825016 to Z.C.), Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, and Tsinghua-Peking Joint Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
J.Y. prepared the sample and performed the biochemical analysis. W.Z. did the initial tries. K.C. performed the EM analysis. Z.C. wrote the manuscript with help from all authors. Z.C. directed and supervised all of the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Blaine Bartholomew, Karl-Peter Hopfner and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CryoEM analysis of the PBAF-nucleosome complex.
(a) A representative negative staining micrograph from 20 micrographs. (b) A representative cryo-EM micrograph from 19,313 micrographs. (c) 2D class averages of characteristic projection views of cryo-EM particles. (The diameter of the circular mask: 32nm.). (d) Flowchart of the cryo-EM data processing. (e) Angular distributions of the cryo-EM particles in the final round of refinement. (f) Resolution estimation of the EM maps. Gold standard Fourier shell correlation (FSC) curves, showing the overall nominal resolutions of 3.4 Å, 2.8 Å, 3.2 Å, 3. 2 Å for the overall complex, motor-nucleosome region, the left part (NBL-DBL) and the right part (NBL-HBL) of the SRM, respectively. (g). Model-map FSC plot calculated by Phenix between the map and the model.
Extended Data Fig. 2 Local density maps of the PBAF-nucleosome complex.
(a) Local cryoEM maps of the motor module at a resolution of 2.8 Å. (b) Local cryoEM maps of the SRM at a resolution of 3.2 Å.
Extended Data Fig. 3 Inter-molecular interactions of the PBAF-nucleosome complex detected by CL-MS.
Subunits are colored as in Fig. 1. Cross-links supported by spatial proximity in the cryoEM structure are shown.
Extended Data Fig. 4 Exit DNA unwrapping and AD binding to NBL.
(a) Local cryoEM density of the nucleosomal DNA bound by the PBAF complex, superimposed with the DNA from the “601” nucleosome (colored grey, PDB code 3MVD)65. (b) Binding of the AD to PHF10 and SMARCB1 in the NBL. The circled region is enlarged in (c). (c) Binding of the AD to the CTL of PHF10. Multiple-sequence alignments of Arid2 proteins and human ARID1a around the binding interface. Conserved residues are highlighted in yellow. (d) Two views of the nucleosome together with the NBL, superimposed with the DNA from the “601” nucleosome (colored grey). The electrostatics of AD of ARID2 is calculated with Pymol.
Extended Data Fig. 5 Structural comparison of the motor domains bound to the nucleosome.
(a) Comparison of the motors of PBAF (colored as Fig. 2) and recombinant BAF (colored blue, PDB code 6ltj)19 bound to the nucleosome. The histone octamers are aligned. (b) Comparison of the motors of PBAF and endogenous BAF (colored pink, PBDDEV_00000056)18 bound to the nucleosome. The histone octamers are aligned. (c-d) Structural comparisons of the motor domains of PBAF (lobe 1 in green, lobe 2 in cyan), to the recombinant BAF (d, blue, PDB code 6ltj)19, endogenous BAF (e, colored pink, PBDDEV_00000056)18. The structures of lobe 1 are aligned. The boxed regions (the Brace helices and the ATPase pocket) are enlarged for further analysis at the bottom. (e) Comparison of the SMARCA4 motor of the PBAF complex and Snf2 motor bound to the nucleosome (colored yellow, PDB code 5Z3U)33. The histone octamers are aligned. (f) Structural comparisons of the motor domains of PBAF and Snf2 (colored yellow, PDB code 5Z3U). The structures of lobe 1 are aligned. The boxed regions are enlarged for further analyses in g and h.
Extended Data Fig. 6 Additional biochemical analyses.
(a) Representative SDS-PAGE gels of the WT and mutant PBAF complexes from 5 independent experiments. (b) Chromatin remodeling activity of WT and R1372A mutant at a substaturating concentration of enzyme. The nucleosome and the enzyme complex at 10 nM and 5 nM were used, respectively. Representative gel is shown on the top, and quantification of the remodeled product at the bottom. Data are presented as mean values +/− SD (n = 3 technical replicates). (c) Chromatin remodeling activities of different batch preparations of the WT and mutant PBAF complex. Nucleosomes at 10 nM, and the enzyme complex at the indicated concentrations were used. Representative gels are shown on the left, and quantification on the right. Data are presented as mean values +/− SD (n = 3 technical replicates).
Extended Data Fig. 7 Local structural analysis of PBAF.
(a) Structural comparison of the NBL of PBAF (color coded) and BAF (grey, PBD code 6ltj)19. SMARCB1 is aligned. (b) Nucleosome binding by the FH of PBAF (left), and BAF (right). (c) Structural comparison of the structural core of the HBL of PBAF (color coded) and BAF (grey). The structures of NTD of SMARCA4 are aligned. (d) Structural comparison of the helical bundle of PBAF (color coded) and BAF (grey). The structures of SMARCD1 are aligned. (e) Structural comparison of ARM of ARID2 (colored gold) and ARID1a (colored grey). The HSA helix is partially melted in PBAF, with Arg448 located at the loop region, whereas it is a part of the helix in BAF. (f) Structural comparison of the ARM domains of ARID2 (colored gold) and Rsc9 in RSC (colored grey, PDB code 6K15)25. (g) Structural comparison of DBL of PBAF (color coded) and RSC (grey) around A2BM region. The structures of A2BM and R6BD (Rsc6-binding domain) are aligned. (h) Structural comparison of the structural core of the HBL of PBAF (color coded) and RSC (grey). The structures of the SANT domain of SMARCC and Rsc8 are aligned.
Extended Data Fig. 8 Multiple sequence alignments of ARID2-like proteins.
The AD domain of human ARID1a is included. The conserved residues involved in binding to the PHF10 are highlighted in yellow.
Extended Data Fig. 9 Multiple sequence alignments of PHF10-like proteins.
The conserved residues of CTL involved in binding to the AD domain of ARID2 are highlighted in yellow.
Extended Data Fig. 10 Multiple sequence alignments of PBRM1-like proteins.
Only the sequences of the CTD are shown. The structures of the N-terminal BD, BAH and HMG domains are not resolved in current study, and not included in the alignments.
Supplementary information
Supplementary Information
This file contains raw data for Fig. 2f and Supplementary Fig. 6a–c.
Rights and permissions
About this article
Cite this article
Yuan, J., Chen, K., Zhang, W. et al. Structure of human chromatin-remodelling PBAF complex bound to a nucleosome. Nature 605, 166–171 (2022). https://doi.org/10.1038/s41586-022-04658-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04658-5
This article is cited by
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking
Nature Communications (2024)
-
Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes
Nature Structural & Molecular Biology (2024)
-
Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties
Communications Biology (2024)
-
ARID3a from the ARID family: structure, role in autoimmune diseases and drug discovery
Acta Pharmacologica Sinica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.