Abstract
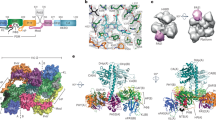
Many aspects of plant photoperception are mediated by the phytochrome (Phy) family of bilin-containing photoreceptors that reversibly interconvert between inactive Pr and active Pfr conformers1,2. Despite extensive biochemical studies, full understanding of plant Phy signalling has remained unclear due to the absence of relevant 3D models. Here we report a cryo-electron microscopy structure of Arabidopsis PhyB in the Pr state that reveals a topologically complex dimeric organization that is substantially distinct from its prokaryotic relatives. Instead of an anticipated parallel architecture, the C-terminal histidine-kinase-related domains (HKRDs) associate head-to-head, whereas the N-terminal photosensory regions associate head-to-tail to form a parallelogram-shaped platform with near two-fold symmetry. The platform is internally linked by the second of two internal Per/Arnt/Sim domains that binds to the photosensory module of the opposing protomer and a preceding ‘modulator’ loop that assembles tightly with the photosensory module of its own protomer. Both connections accelerate the thermal reversion of Pfr back to Pr, consistent with an inverse relationship between dimer assembly and Pfr stability. Lopsided contacts between the HKRDs and the platform create profound asymmetry to PhyB that might imbue distinct signalling potentials to the protomers. We propose that this unique structural dynamism creates an extensive photostate-sensitive surface for conformation-dependent interactions between plant Phy photoreceptors and their signalling partners.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Full versions of all SDS–PAGE gels and blots are provided in Supplementary Fig. 1. The 3D cryo-EM map of the full-length Arabidopsis PhyB at 3.3 Å resolution has been deposited in the Electron Microscopy Data Bank database under accession code EMD-24780. The corresponding atomic model has been deposited in the RCSB Protein Data Bank under accession code 7RZW. This study made use of several publicly available protein structures obtained from the RCSB Protein Data Bank (http://www.rcsb.org) under accession codes 4OUR, 6TC5, 3DGE, 4GCZ, 4U7O and 4I5S. Source data are provided with this paper.
References
Legris, M., Ince, Y. C. & Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10, 5219 (2019).
Burgie, E. S. & Vierstra, R. D. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 (2014).
Auldridge, M. E. & Forest, K. T. Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 (2011).
Rockwell, N. C., Su, Y. S. & Lagarias, J. C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006).
Franklin, K. A. & Quail, P. H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 (2010).
Jung, J. H. et al. Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016).
Legris, M. et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016).
Burgie, E. S. et al. Differing biophysical properties underpin the unique signaling potentials within the plant phytochrome families. Proc. Natl Acad. Sci. USA 118, e2105649118 (2021).
Essen, L. O., Mailliet, J. & Hughes, J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl Acad. Sci. USA 105, 14709–14714 (2008).
Wagner, J. R., Brunzelle, J. S., Forest, K. T. & Vierstra, R. D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005).
Yang, X., Kuk, J. & Moffat, K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl Acad. Sci. USA 105, 14715–14720 (2008).
Takala, H. et al. Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014).
Burgie, E. S., Zhang, J. & Vierstra, R. D. Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016).
Burgie, E. S. et al. Photoreversible interconversion of a phytochrome photosensory module in the crystalline state. Proc. Natl Acad. Sci. USA 117, 300–307 (2020).
Isaksson, L. et al. Signaling mechanism of phytochromes in solution. Structure 29, 151–160 (2021).
Anders, K., Daminelli-Widany, G., Mroginski, M. A., von Stetten, D. & Essen, L. O. Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 (2013).
Bhoo, S. H., Davis, S. J., Walker, J., Karniol, B. & Vierstra, R. D. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414, 776–779 (2001).
Yeh, K. C., Wu, S. H., Murphy, J. T. & Lagarias, J. C. A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508 (1997).
Li, F. W. et al. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 6, 7852 (2015).
Rockwell, N. C. & Lagarias, J. C. Phytochrome evolution in 3D: deletion, duplication, and diversification. New Phytol. 225, 2283–2300 (2020).
Yeh, K. C. & Lagarias, J. C. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl Acad. Sci. USA 95, 13976–13981 (1998).
Boylan, M. T. & Quail, P. H. Are the phytochromes protein kinases? Protoplasma 195, 12–17 (1996).
Elich, T. D. & Chory, J. Phytochrome: if it looks and smells like a histidine kinase, is it a histidine kinase? Cell 91, 713–716 (1997).
Ni, W. et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164 (2014).
Buckley, C. E. et al. Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev. Cell 36, 117–126 (2016).
Chernov, K. G., Redchuk, T. A., Omelina, E. S. & Verkhusha, V. V. Near-infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 117, 6423–6446 (2017).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Shimizu-Sato, S., Huq, E., Tepperman, J. M. & Quail, P. H. A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044 (2002).
Krall, L. & Reed, J. W. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl Acad. Sci. USA 97, 8169–8174 (2000).
Matsushita, T., Mochizuki, N. & Nagatani, A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574 (2003).
Burgie, E. S. et al. Photosensing and thermosensing by phytochrome B require both proximal and distal allosteric features within the dimeric photoreceptor. Sci. Rep. 7, 13648 (2017).
Burgie, E. S., Bussell, A. N., Walker, J. M., Dubiel, K. & Vierstra, R. D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl Acad. Sci. USA 111, 10179–10184 (2014).
Nagano, S. et al. Structural insights into photoactivation and signalling in plant phytochromes. Nat. Plants 6, 581–588 (2020).
Diensthuber, R. P., Bommer, M., Gleichmann, T. & Moglich, A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 (2013).
Wang, C. et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 11, e1001493 (2013).
Cai, Y. et al. Conformational dynamics of the essential sensor histidine kinase WalK. Acta Crystallogr. D 73, 793–803 (2017).
Casino, P., Rubio, V. & Marina, A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009).
Lagarias, J. C. & Mercurio, F. M. Structure function studies on phytochrome. Identification of light-induced conformational changes in 124-kDa Avena phytochrome in vitro. J. Biol. Chem. 260, 2415–2423 (1985).
Jones, A. M., Vierstra, R. D., Daniels, S. M. & Quail, P. The role of separate molecular domains in the structure of phytochrome from etiolated Avena sativa L. Planta 164, 501–516 (1985).
Li, H., Zhang, J., Vierstra, R. D. & Li, H. Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl Acad. Sci. USA 107, 10872–10877 (2010).
Gourinchas, G. et al. Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci. Adv. 3, e1602498 (2017).
Etzl, S., Lindner, R., Nelson, M. D. & Winkler, A. Structure-guided design and functional characterization of an artificial red light-regulated guanylate/adenylate cyclase for optogenetic applications. J. Biol. Chem. 293, 9078–9089 (2018).
Mechaly, A. E., Sassoon, N., Betton, J. M. & Alzari, P. M. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 12, e1001776 (2014).
Shin, A. Y. et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7, 11545 (2016).
Klose, C., Nagy, F. & Schafer, E. Thermal reversion of plant phytochromes. Mol. Plant 13, 386–397 (2020).
Klose, C. et al. Systematic analysis of how phytochrome B dimerization determines its specificity. Nat. Plants 1, 15090 (2015).
Rensing, S. A., Sheerin, D. J. & Hiltbrunner, A. Phytochromes: more than meets the eye. Trends Plant Sci. 21, 543–546 (2016).
Kikis, E. A., Oka, Y., Hudson, M. E., Nagatani, A. & Quail, P. H. Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 5, e1000352 (2009).
Ni, W. et al. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017).
Acknowledgements
Cryo-EM data were collected using the Titan Krios system at the David Van Andel Cryo-Electron Microscopy Suite at the Van Andel Institute. We thank G. Zhao and X. Meng for help with data collection; J. Zhang for the mass spectrometry analysis; H. Zaher for help with the kinase assays; and C. Sherman and K. McLoughlin for technical assistance. This work was funded by the US National Institutes of Health R01 grants GM127892 (to R.D.V.) and GM131754 (to Huilin Li), and funds provided by the Van Andel Institute (to Huilin Li) and Washington University in St Louis (to R.D.V.).
Author information
Authors and Affiliations
Contributions
Hua Li, E.S.B., Huilin Li, Z.T.K.G. and R.D.V. designed the experiments. Hua Li performed the cryo-EM and 3D reconstruction. Hua Li and E.S.B. built and refined the atomic models. E.S.B. and Z.T.K.G. performed the mutagenesis and spectroscopy assays. Z.T.K.G. conducted the limited proteolysis and kinase assays. Hua Li, E.S.B., Huilin Li and R.D.V. wrote the manuscript with input from Z.T.K.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Jorge Casal, Elizabeth Getzoff and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Workflow and resolution estimation for the cryo-EM map of Arabidopsis PhyB.
a, SDS-PAGE analysis of the recombinant full-length biliprotein. Gels were either stained for protein with Coomassie blue (left) or assayed for bound PΦB by zinc-induced fluorescence (right). MM, molecular mass standards. Samples were indistinguishable to those described by Burgie et al.2 b, UV-vis absorbance spectra of PhyB. The spectra were collected from dark-adapted samples (Pr) or after saturating irradiation with 660-nm red light (RL, mostly Pfr). Absorption maxima were determined from the difference spectrum shown at 70% amplitude. The spectral change ratio (SCR)8 at 723 nm is indicated in parenthesis. c, Work flow for data processing of the cryo-EM images of the PhyB dimer. In the first refined overall map at 3.4-Å resolution, all PhyB domains were present but the regions encompassing the PAS1 domain were poorly resolved. Focused refinement, excluding the PAS1 domains, led to the 3.3-Å final map (lower left panel). Lower right panel shows that the EM density of the flexible PAS1 domains (purple), which were captured at 15-Å resolution by 3D variability analysis of down-sampled particle images. d–h, Resolution estimation of the 3.3-Å 3D map. d, A representative cryo-EM micrograph sampled from 6,153 micrographs collected. e, Sampling of 2D class averages. f, Colored-coded local resolution of the 3D map. g, Eulerian angle distribution of raw particle images used in the final 3D reconstruction. h, Gold-standard Fourier shell correlation (FSC) and the validation of the atomic model by correlation curves comparing the model to the final and two half maps.
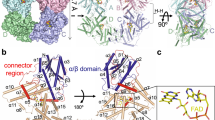
Extended Data Fig. 2 Superposition of the cryo-EM map densities of the PSM, PΦB, and the bilin-binding GAF domain pocket with the X-ray crystallographic model of the PSM.
Motifs/residues are colored as in Fig. 1b. a, Fitting of the nPAS, GAF and PHY domains, and the hairpin (HP) motif within the EM map density (light grey surface) of protomer A and protomer B. PΦB is shown in red. b, Superposition of the PhyB PSM determined by cryo-EM of the full-length PhyB (protomer A; slate blue) and by X-ray crystallography of the PhyB PSM (grey; residues 90-624, PDB ID code, 4OUR32). c, PΦB conformations (in sticks) in protomers A and B modeled within the EM map density (grey mesh). The A-D pyrrole rings are labeled along with Cys357 that forms a thioether linkage to the C31 carbon of PΦB. The D ring C182 carbon is indicated. d, Superposition of the PΦB structures determined by cryo-EM of the full-length PhyB protomer A (colored) and by X-ray crystallography of the PhyB PSM (grey). e, The bilin-binding pockets of protomers A and B highlighting neighboring amino acids (sticks) and superposed in the EM map density (grey mesh). f, Superposition of the bilin-binding pocket determined from the cryo-EM structure of full-length PhyB protomer A with the X-ray crystallographic structure of the PhyB PSM.
Extended Data Fig. 3 Superposition of the cryo-EM map densities of various PhyB motifs with the cryo-EM model.
Shown in sticks are various amino acids modeled within the EM densities (grey mesh). Domains/residues are colored as in Fig. 1b. a–c, The NTE and knot lasso region of protomer A showing the knot lasso (a) and NTE separately (b), and combined (c). d, The hairpin loop extending from the PHY domain in protomer A. e, Portions of the DHp regions within the HKRD from protomers A and B. Gln937, which positionally corresponds to the conserved phosphoacceptor histidine found in prokaryotic transmitter histidine kinases, is circled in red. f, The modulator loop extending from between the PAS1 and PAS2 domains within protomer A. g, Residues within the CA domain of the HKRD in protomer A surrounding a possible ATP-binding pocket.
Extended Data Fig. 4 Topology of PhyB generated from the cryo-EM 3D model and structural predictions of the three PAS domains.
a, Topological schematic of PhyB. Shown are the NTE, nPAS, GAF, PHY and PAS2 domains, and the DHp and CA domains within the HKRD highlighting the positions, lengths, and contacts of the α-helices and β-strands, and the contacts for the knot lasso, hairpin, and modulator loop features. The position of PΦB within the GAF domain β-sheet is indicated. The entry and exit points of the poorly resolved PAS1 domain between the PHY and PAS2 domains are shown. The GAF and nPAS loops unique to plant Phys and the PHY domain hairpin residues (WGG and PRXSF) involved in a predicted β-strand to α-helical transformation during photoconversion are identified in the ellipsoids32,33. The predicted ATP-binding region is highlighted by the yellow ovals. The H1a cruciate region within the helix α1 of the DHp, which provides the head-to-tail to head-to-head crossover point with Cys925 at its center, is located by the dark blue box. Gln937 is the residue that replaces the phosphoacceptor histidine found in prokaryotic two-component HKs. Amino acid sequence conservation of each feature can be found in Extended Data Fig. 6. b–d, Structural predictions of the three PAS domains in PhyB using TrRosetta62. The PAS domain cores are circled by the dashed grey line, which is followed by an α/β roll64. Terminal amino acids are indicated. b and c, Superposition of 3D models of the nPAS and PAS2 domains determined by cryo-EM with those predicted (p) by TrRosetta. The cartoons on the left are the cryo-EM models and those on the right are superpositions of the models (grey) with those calculated (rainbow) (rmsd = 1.1 Å for nPAS and 1.0 Å for PAS2). d, Predicted models of the PAS1 region by TrRosetta. The left cartoon is a prediction for the PAS1 domain plus 31 additional N-terminal residues not found in the cryo-EM model. The middle cartoon includes the top five predictions for the PAS1 domain alone. The right cartoon is a superposition of the predicted PAS1 domain with the PAS2 cryo-EM model (rmsd = 1.3 Å).
Extended Data Fig. 5 Limited protease sensitivity is consistent with the cryo-EM model of PhyB.
a, Concentration-dependent cleavage of PhyB by chymotrypsin and GluC. Purified full-length PhyB was incubated for 15 min with increasing amounts of each protease and then subjected to complete digestion with trypsin followed by tandem MS identification of peptides generated by each protease. Peptides that ended in chymotrypsin or GluC cut sites were quantified from the MS1 scans. Each row represents a potential cleavage site; white bars indicate no cleavage whereas green boxes represent regions without detectable peptides. All MS data represent the means of four technical replicates. The digestions were aligned with the domain architecture of PhyB (see Fig. 1a). b, Relative susceptibility to proteolysis for all cleaved sites at 2 ng/µL chymotrypsin or GluC. Bars are colored by domain as in (a). c, Proportion of cleavage sites within each domain that were either susceptible to high or low concentrations of both proteases, not cleaved, or not detected. d–f, 3D views of the protease-sensitive sites in PhyB highlighting: (d) the NTE-nPAS-GAF-PHY-PAS2, (e) HKRD, and (f) PSM regions. The PhyB structure is shown in cartoon while the cleavage sites are highlighted in spheres and color-coded based on protease sensitivity. Protomers A and B are presented in grey and white, respectively. Residues involved in dimerization are highlighted in magenta in (f).
Extended Data Fig. 6 Amino acid sequence alignment of the PhyA, PhyB, PhyC and PhyE subfamilies within angiosperms.
See ref. 8 for full description of the sequence list. The font height of each amino acid is proportional to its percent homology within each Phy isoform subfamily. The positions of the NTE, nPAS, GAF, PHY, PAS1, PAS2, and DHp and CA domains of the HKRD are located by the red, blue, green, orange, gray, magenta, brown and cyan bars, respectively. The PAS and GAF loops, the knot lasso, and the hairpin and modulator features are located by light blue, light green, turquoise, yellow, and dark red bars, respectively. The α-helices and β-strands, along with their numbering within each domain, are shown below the sequence by the coiled and wavy lines, respectively. The red star locates the position that commonly contains the phosphoacceptor histidine within prokaryotic transmitter HK domains. The blue circles indicate the core amino acids within the N-terminal knot. Red arrows locate the end point for the six N-terminal truncations of PhyB analyzed in this study (N624, N778, N799, N908, N928, and N982). Green arrowheads locate the residues shown experimentally to promote GAF-PAS2 contacts within the dimer. Red diamonds locate residues predicted to form the ATP-binding pocket in the CA domain based on prokaryotic transmitter HKs. A green circle locates Cys925 that is at the center of the cruciate crossover that transitions the PhyB protomers from head-to-tail to head-to-head arrangements.
Extended Data Fig. 7 Structural and enzymatic analyses of PhyB reveal its homology to transmitter HKs but with a compromised phosphotransferase activity.
a, Cartoon 3D structure of the paired HKRDs from Arabidopsis (At) PhyB showing the structures and inter-molecular interfaces between the CA and DHp domains. Images on the right show a pair of orthlogonal views with residues within one half of the HKRD dimer interface shown as spheres. These residues were contributed by helix α1 from the DHp of protomer B, and helix α2 of DHp and helices α1, α2 and α4 from the CA domain of protomer A. b, The network of intermolecular contacts between the DHp and CA domains in (a) illustrated for simplicity. c, Top views of the DHp regions of the HKRDs for At PhyB as compared to the same region in the prokaryotic HK853 transmitter HK from Thermotoga maritima (PDB ID 3DGE37). Gln937 in At PhyB and the phosphoacceptor histidine in Tm HK853 are shown in red sticks. d, Closeup 3D views of the DHp domains in At PhyB corresponding to the region surrounding phosphoacceptor histidine in transmitter HKs. Gln937 in PhyB, which is a histidine in transmitter kinases, is circled. e, 3D superposition of the CA domain in At PhyB shown in cartoon with those from several bacterial two-component HKs illustrating its HK ancestry. Representatives include YF1 from Bacillus subtilis (Bs) (PDB ID 4GCZ), WalK from Lactobacillis plantarum (Lp) (PDB ID 4U7O), HK853 from Thermotoga maritima (Tm) (PDB ID 3DGE), and Vick from Streptococcus mutans (Sm) (PDB ID 4I5S). f, Model showing the predicted position of ADP (red) in the AtPhyB CA domain when modeled after that for LpWalK. Residues that might participate in binding are indicated. ADP clashes with multiple residues in the pocket of this predicted AtPhyB model, suggesting that conformational shifts in AtPhyB induced by ATP or upon photoactivation would be necessary for binding. g, Schematic of binding interactions between the ADP analogue adenylyl-imidodiphosphate (AMPPNP) and CA domain from the Lp WalK determined by X-ray crystallography (left; PDB ID 4U7O36) and that predicted for AtPhyB when modelled after the LpWalK structure (right). Hydrogen bonds and representative hydrophobic interactions are indicated with green and red dashed lines, respectively. Analogous residues are depicted in similar positions in schematics, except for LpAsn514 and AtSer1054. h and i, AtPhyB is a poor protein kinase as compared to Pseudomonas syringae (Ps) BphP based on autophosphorylation assays. The recombinant biliproteins were incubated at ambient temperature (~24°C) with 150 μM ATP supplemented with 10 μCi of [γ-32P]-ATP, quenched with SDS-PAGE sample buffer, and subjected to SDS-PAGE. Shown are the SDS-PAGE gels assayed for bound 32P by autoradiography or stained for protein with Coomassie blue. h, Time course for autophosphorylation of PsBphP as Pfr. i, Comparisons of autophosphorylation activities of AtPhyB as Pr and Pfr with those of PsBphP. Reactions containing equal mass amounts of biliprotein were terminated after 2 hr. (left) Autoradiography of the kinase reactions. (right) SDS-PAGE gel showing the biliprotein preparations used. Arrowheads locate PsBphP. The phosporimager scans are representative of 3 independent experiments. Full gels can be found in Supplementary Fig. 1.
Extended Data Fig. 8 SDS-PAGE analyses and absorption spectra of the PhyB truncations and point mutations studied here.
a and d, UV-vis absorbance spectra. The absorption spectra were collected from dark-adapted samples (Pr) or after saturating irradiation with 660-nm red light (RL, mostly Pfr). Absorption maxima were determined from the difference spectrum shown at 70% amplitude. SCR values are indicated in parentheses. Spectra represent the mean of three technical replicates. b and c, SDS-PAGE analysis of the purified PhyB proteins described in (a) and (d). After electrophoresis, the biliproteins were either stained for protein with Coomassie blue or imaged for the bound PΦB by zinc-induced fluorescence under UV light. Full gels can be found in Supplementary Fig. 1.
Supplementary information
Supplementary Information
Supplementary Methods and the accompanying references, and the legends for Supplementary Fig. 1, Supplementary Videos 1 and 2 and the source data.
Supplementary Video 1
Cryo-EM structure of Arabidopsis PhyB
Supplementary Video 1
Morph between protomers A and B of Arabidopsis PhyB
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, H., Burgie, E.S., Gannam, Z.T.K. et al. Plant phytochrome B is an asymmetric dimer with unique signalling potential. Nature 604, 127–133 (2022). https://doi.org/10.1038/s41586-022-04529-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04529-z
This article is cited by
-
Structural insights into plant phytochrome A as a highly sensitized photoreceptor
Cell Research (2023)
-
Plant phytochrome A in the Pr state assembles as an asymmetric dimer
Cell Research (2023)
-
The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms
Nature Plants (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.