Abstract
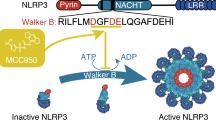
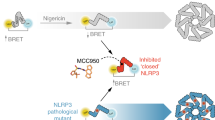
NLRP3 is an intracellular sensor protein that when activated by a broad spectrum of exogenous and endogenous stimuli leads to inflammasome formation and pyroptosis1,2. The conformational states of NLRP3 and the way antagonistic small molecules act at the molecular level remain poorly understood2,3. Here we report the cryo-electron microscopy structures of full-length human NLRP3 in its native form and complexed with the inhibitor CRID3 (also named MCC950)4. Inactive, ADP-bound NLRP3 is a decamer composed of homodimers of intertwined leucine-rich repeat (LRR) domains that assemble back-to-back as pentamers. The NACHT domain is located at the apical axis of this spherical structure. One pyrin domain dimer is in addition formed inside the LRR cage. Molecular contacts between the concave sites of two opposing LRR domains are mediated by an acidic loop that extends from an LRR transition segment. Binding of CRID3 considerably stabilizes the NACHT and LRR domains relative to each other. CRID3 binds into a cleft, connecting four subdomains of the NACHT with the transition LRR. Its central sulfonylurea group interacts with the Walker A motif of the NLRP3 nucleotide-binding domain and is sandwiched between two arginine residues, which explains the specificity of NLRP3 for this chemical entity. With the determination of the binding site of this key therapeutic agent, specific targeting of NLRP3 for the treatment of autoinflammatory and autoimmune diseases and rational drug optimization is within reach.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density reconstructions and models have been deposited in the Electron Microscopy Data Bank (EMDB) (accession codes EMD-13687 for the NLRP3 apo decamer and EMD-13684, EMD-13685, EMD-13686, EMD-13692, EMD-13693 and EMD-13699 for the NLRP3–ADP–CRID3 decamer), and in the PDB under accession code 7PZC. All data are available in the Article or its Supplementary Information files. Source data are provided with this paper.
References
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Swanson, K. V., Deng, M. & Ting, J. P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Latz, E., Xiao, T. S. & Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 (2013).
Laliberte, R. E. et al. Glutathione S-transferase omega 1-1 is a target of cytokine release inhibitory drugs and may be responsible for their effect on interleukin-1β posttranslational processing. J. Biol. Chem. 278, 16567–16578 (2003).
Song, N. et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 68, 185–197 (2017).
Schmid-Burgk, J. L. et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103–109 (2016).
Shi, H. et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016).
He, Y., Zeng, M. Y., Yang, D., Motro, B. & Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016).
Mangan, M. S. J. et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17, 588–606 (2018).
Primiano, M. J. et al. Efficacy and pharmacology of the NLRP3 inflammasome inhibitor CP-456,773 (CRID3) in murine models of dermal and pulmonary inflammation. J. Immunol. 197, 2421–2433 (2016).
Harrison, D. et al. Discovery of a series of ester-substituted NLRP3 inflammasome inhibitors. Bioorg. Med. Chem. Lett. 30, 127560 (2020).
Coll, R. C. et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015).
Vande Walle, L. et al. MCC950/CRID3 potently targets the NACHT domain of wild-type NLRP3 but not disease-associated mutants for inflammasome inhibition. PLoS Biol. 17, e3000354 (2019).
Corcoran, S. E., Halai, R. & Cooper, M. A. Pharmacological inhibition of the Nod-like receptor family pyrin domain containing 3 inflammasome with MCC950. Pharmacol. Rev. 73, 968–1000 (2021).
Sharif, H. et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019).
Kobe, B. & Deisenhofer, J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J. Mol. Biol. 264, 1028–1043 (1996).
Bae, J. Y. & Park, H. H. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J. Biol. Chem. 286, 39528–39536 (2011).
Maekawa, S., Ohto, U., Shibata, T., Miyake, K. & Shimizu, T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 7, 11813 (2016).
Hu, Z. et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013).
Tenthorey, J. L. et al. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888–893 (2017).
Erzberger, J. P. & Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006).
Hoss, F. et al. Alternative splicing regulates stochastic NLRP3 activity. Nat. Commun. 10, 3238 (2019).
Touitou, I. et al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum. Mutat. 24, 194–198 (2004).
Coll, R. C. et al. MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 15, 556–559 (2019).
Tapia-Abellán, A. et al. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 15, 560–564 (2019).
Wendler, P., Ciniawsky, S., Kock, M. & Kube, S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823, 2–14 (2012).
Compan, V. et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487–500 (2012).
Zhang, Z. et al. Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J. Exp. Med. 214, 2671–2693 (2017).
Green, J. P. et al. Chloride regulates dynamic NLRP3-dependent ASC oligomerization and inflammasome priming. Proc. Natl Acad. Sci. USA 115, E9371–E9380 (2018).
Hafner-Bratkovič, I. et al. NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway. Nat. Commun. 9, 5182 (2018).
Hochheiser, I. V. et al. Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation. Preprint at https://doi.org/10.1101/2021.11.25.470035 (2021).
Andreeva, L. et al. Full-length NLRP3 forms oligomeric cages to mediate NLRP3 sensing and activation. Preprint at https://doi.org/10.1101/2021.09.12.459968 (2021).
Schwaid, A. G. & Spencer, K. B. Strategies for targeting the NLRP3 inflammasome in the clinical and preclinical space. J. Med. Chem. 64, 101–122 (2021).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1055–1064 (2020).
Dekker, C. et al. Crystal structure of NLRP3 NACHT domain with an inhibitor defines mechanism of inflammasome inhibition. J. Mol. Biol. 433, 167309 (2021).
Hochheiser, I. V. et al. Cryo-EM structure of the NLRP3 decamer bound to the cytokine release inhibitory drug CRID3. Preprint at https://doi.org/10.1101/2021.07.22.453353 (2021).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Prisant, M. G., Williams, C. J., Chen, V. B., Richardson, J. S. & Richardson, D. C. New tools in MolProbity validation: CaBLAM for cryoEM backbone, UnDowser to rethink “waters,” and NGL Viewer to recapture online 3D graphics. Protein Sci. 29, 315–329 (2020).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Keuler, T. et al. Development of fluorescent and biotin probes targeting NLRP3. Front. Chem. 9, 642273 (2021).
Kofoed, E. M. & Vance, R. E. Blue native polyacrylamide gel electrophoresis to monitor inflammasome assembly and composition. Methods Mol. Biol. 1040, 169–183 (2013).
Sester, D. P. et al. A novel flow cytometric method to assess inflammasome formation. J. Immunol. 194, 455–462 (2015).
Zhang, L. et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015).
Park, K. et al. Control of repeat-protein curvature by computational protein design. Nat. Struct. Mol. Biol. 22, 167–174 (2015).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
Access to cryo-EM time was supported by iNEXT-Discovery grant PID 11967 for the NLRP3–CRID3 sample and INSTRUCT grant PID 11340 for the NLRP3 apo sample. We thank W. Hagen, F. Weis and A. Durand for cryo-EM data recordings; H. Stark for initial advice on electron microscopy sample preparation; and A. Steiner for instructions on cell assays. C.E. thanks B. Böttcher for facility access, R. Witzgall for JEM2100F access, G. Meister for shared infrastructure, G. Lehmann and N. Eichner for IT support and all members of the Structural Biochemistry Group of the Regensburg Center for Biochemistry for help and support. C.E. and M.P. acknowledge funding by the Emmy Noether Programme (DFG grant EN 1204/1-1 to C.E.) and SFB 960 (TP-A8 to C.E.). M.G. and E.L. are funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy–EXC2151–390873048.
Author information
Authors and Affiliations
Contributions
I.V.H. performed all experiments and data analyses, except for the following. J.M. characterized the interface mutants in cells. M.M. performed SPR experiments. R.B. identified peak 2. I.V.H. and M.P. prepared cryo-EM samples. I.V.H., M.P., G.H. and C.E. processed cryo-EM data. G.H., M.G., M.P. and C.E. built the model. I.V.H., M.P., G.H. C.E. and M.G. interpreted the data. E.L. provided reagents for cell-based assays, and contributed to discussions. C.E. supervised cryo-EM sample preparation, optimization and data interpretation and contributed to funding. M.G. designed and supervised research and wrote the manuscript. All authors discussed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.G. and E.L. are co-founders and consultants of IFM Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Preparation of full length, wild-type human NLRP3 for electron microscopy analyses.
a, Analytical SEC of recombinant, human MBP–NLRP3 (3–1036) reveals two elution peaks, one close to the void volume (peak 1) and one at a size larger than 1 MDa (peak 2). The peak ratio is shifted from peak 1 under standard buffer conditions (50 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM ADP, 0.5 mM TCEP) towards peak 2 by addition of CRID3 (10 µM CRID3). Analytical SEC runs were performed on a Superose 6 Increase 10/300 GL column with 700 µl applied at a concentration of 3 mg/ml (repeated >3 times). b, Coomassie-stained SDS PAGE analysis of purified MBP-NLRP3 (repeated >3 times). c, SEC–MALS experiment of MBP-NLRP3 Peak 2 plus ADP on a Superose 6 Increase 10/300 GL column. The calculated mass of an MBP-NLRP3 monomer is 158 kDa. d, SEC–MALS experiment of MBP-NLRP3 Peak 2 plus ADP plus CRID3 on a Superose 6 Increase 10/300 GL column. The average apparent mass of Peak 2 from both experiments equals 10-times the calculated mass of the protein. e, Analytical SEC of recombinant, human NLRP3 (3–1036) after TEV digestion using a Superose 6 Increase 3.2/300 column. f, Negative-stain micrograph of TEV-digested, gel filtered NLRP3 plus CRID3 plus BS3 Peak 2 elution fraction that was used for subsequent cryo-EM grid preparation (right, one among a few hundred images). An SDS-PAGE analysis of the protein w/o BS3 is shown left. The scale-bar equals 200 nm. g, Exposure of NLRP3 peak 2 to ATP does not convert the decamer assembly into another state. MBP-NLRP3 expressed in Sf9 cells and purified by affinity chromatography in the absence of any nucleotide elutes in two peaks (top). Incubation of MBP-NLRP3 peak 2 (from top) with 1 mM ATP•Mg2+ for 2 h at 4 °C does not lead to a change in the elution profile, indicating the autoinhibition of the decamer assembly (bottom). h, THP-1 cells contain an oligomeric endogenous NLRP3 species. Immunoblotting of BN-PAGE of THP-1 cell lysate supernatant and pellet fractions from PMA-differentiated, untreated and LPS-treated wild-type cells pre-incubated with or without CRID3 (repeated 2 times). The supernatant of LPS-treated and BS3 cross-linked cell lysates contains an oligomeric NLRP3 fraction in the presence or absence of CRID3 (left). In the pellet fractions of LPS-treated cell lysates only an oligomeric NLRP3 species was detected at a molecular mass around 1.15 MDa, in line with the formation of a decamer (middle). Anti-NLRP3 Cryo-2 and anti-β actin antibodies were used for the WB analysis. BN-PAGE analysis of recombinant human (MBP-)NLRP3 protein expressed in Sf9 cells and purified by affinity chromatography and SEC (right). Source data are provided as a source data file.
Extended Data Fig. 2 Biochemical characterization of NLRP3 peak 1 and peak 2.
a, Analytical SEC of NLRP3 (3–1036) mutants S198E and S198A. The phospho-mimetic S198E mutant shows a decreased content of peak 2. b, Using a multi-cycle turnover reverse-phase HPLC assay with 3 µM protein and 100 µM ATP concentration at 25 °C, NLRP3 peak 1 shows an ~10-times higher ATP turnover number than peak 2. c, Negative stain electron micrographs of TEV-cleaved human NLRP3 peak 1 or peak 2, co-incubated with ASC-mCherry in a 1:10 molar ratio or ASC-mCherry incubated alone for 3 min at 25 °C. All samples show a comparable prevalence of ASC filament formation events. Images are representative of two independent experiments. The scale bars are 500 nm. d, Pull-down experiments of Sf9 cell expressed GST-NEK7 (bait) with MBP-NLRP3 peak 2 (prey) after incubation for 2 h at 4 °C. Experiments were repeated 2 times. e, Sf9 cell expressed GST-NEK7 and MBP-NLRP3 (peak 2) were separately purified and co-incubated (2x molar excess of GST-NEK7) at 4 °C for 4 h and subjected to gel filtration on a Superose 6 increase 10/300 GL column (repeated 3 times). Both recombinant proteins elute separately from each other. f, Elution profile (S6 increase 10/300 GL column) and SDS-PAGE analysis of Sf9 cell co-expressed MBP-NLRP3 and GST-NEK7 (repeated 2 times). An elution peak containing both proteins indicates the formation of a NLRP3–NEK7 heterodimer complex. Source data are provided as a source data file.
Extended Data Fig. 3 Cryo-EM data processing of apo-NLRP3 and the NLRP3–CRID3 complex.
a, Representative cryo-EM micrograph of the human NLRP3 oligomer in the apo state eluting as Peak 2 (replicated more than 35,000 times). Scale bar, 50 nm. b, Representative 2D class averages. c, Processing tree describing particle classification of the NLRP3 cryo-EM data in RELION3.135. d, Top, tilted, and side views of the cryo-EM density of the NLRP3 protein w/o CRID3 at ~10 Å resolution. The 3.9 Å structure of the NLRP3–CRID3 complex is fitted into the density map showing that the apo and the inhibitor-bound NLRP3 decamer structures do not exhibit large conformational rearrangements. e, Representative 2D class averages of the NLRP3–CRID3 complex. f, Processing tree of cryo-EM data in cryo-SPARC leading to 3 ab initio models. g, The second model, which was based on 409,755 particles, was further refined with D5 symmetry resulting in a 3.8 Å resolution map. h, Model 2 was further processed into two 3D classes, one of which contained 237,390 particles. Non-uniform (NU) refinement of the latter class in C1 symmetry led to a 4.8 Å resolution map. The centre region of the NLRP3 cage showed extra density that was fitted with a PYD dimer as described in the main text.
Extended Data Fig. 4 Cryo-EM data processing of the NLRP3–CRID3 complex.
a, Representative cryo-EM micrograph of the human NLRP3 decamer in complex with CRID3 (replicated more than 20,000 times). Scale bar, 50 nm. b, c, Representative 2D class averages. d, e, Processing tree of cryo-EM data in RELION3.135 as described in the Methods section and gold standard FSC plots of masked final maps (grey) as calculated from phenix-mtriage40. f–h, Orientation distribution of particles and heat maps of local resolution estimations.
Extended Data Fig. 5 Quality of cryo-EM densities.
Sections of focused-refined NLRP3–CRID3 cryo-EM density overlaid with their respective atomic models. Densities are shown as a blue mesh, and sticks are shown for the structure model coloured as in Fig. 2a. The labels ‘*’ and ‘†’ refer to the ‘multibody refine monomer’ and ‘best decamer’ density, respectively.
Extended Data Fig. 6 Conformational transitions in NLRP3.
a, Overlay of NLRP3 (7PZC) from the decamer structure with the NLRP3–NEK7 complex structure (6NPY)15. The proteins were aligned to the NBD-HD1 subdomains. Only the last three repeats of the LRRs are coloured for clarity. The C-terminal lobe of NEK7 (blue) adopts the space in the concave site of the LRR that is occupied in the NLRP3 decamer structure by the acidic loop (red). b, Overlay of NLRP3 from the decamer assembly with NLR family proteins NOD2 (5IRN)18, monomeric NLRC4 (4KXF)19, and the disc-like NLRC4 (3JBL)48 structure. c, Amino acid register shift in HD2 and trLRR between the NLRP3 decamer and the NLRP3–NEK7 structure. Overlay of residues 435–828 between our structure (7PZC, dark grey/coloured) and the NLRP3–NEK7 structure (6NPY, light grey)15 showing WHD, HD1, trLRR and 3 repeats of the cnLRR. There is a shift in the amino acid register starting from position E538 that varies between -12 residues in the first helix of HD2 and +44 residues in the first β-strand of trLRR. The register synchronizes again at L737 at the beginning of the cnLRR domain. Segments are coloured according to the amino acid register shifts.
Extended Data Fig. 7 Arrangements of the trLRR and the cnLRR.
a, Sequence alignment of individual repeats of the LRR domain. The trLRR starts at position F650 with the FXXIXI motif and a 26-aa repeat. An LRR-mismatching region of 42 residues interrupts the conventional fold from residue F683 on, forming a flexible, acidic loop. A highly charged stretch of 14 residues (689–702, theoretical pI 4.4) with acidic residues at the tip binds into the concave side of the LRR. The cnLRR starts at position L743 and contains 10 repeats of a proto-typic 28/29 residue alteration49. Charged residues in the concave surface of the LRR are coloured blue and red. Leucine residues or homologous hydrophobic residues at LRR-defining positions are indicated bold, and cysteines are boxed yellow. Mismatching residues that preclude a cnLRR fold are boxed cyan. Secondary structure elements of a cnLRR fold are indicated at the top. b, Electrostatics of the LRR–acidic loop–LRR’ interaction. On the left side is the acidic loop interaction in the LRR without the loop (650–1036, Δ683–727) shown. On the right is the interaction of the C-terminal repeat (998’-1036’) of the cognate LRR binding into the concave LRR side shown.
Extended Data Fig. 8 Mapping of pathogenic disease mutations in human NLRP3.
a, Structural mapping of all missense mutations listed in the Infevers database23. Missense mutations are highlighted as spheres in light blue; validated, pathogenic mutations are marked in red. Although disease mutations are found in the entire protein, a particular accumulation is seen in the NBD of the NACHT domain. b, Overview of the 20 validated CAPS-associated pathogenic mutations from the Infevers database23 with their predicted effect based on the NLRP3 structure. CAPS, Cryopyrin associated periodic syndrome; CINCA, chronic infantile neurological cutaneous articular; FCAS1, familial cold auto-inflammatory syndrome 1; NOMID, neonatal-onset multisystem inflammatory disorder; MWS, Muckle-Wells syndrome. c, Nineteen of twenty pathogenic NLRP3 mutations locate to an interface in the NACHT domain that shears upon activation. The proposed change to an ‘open’ conformation would lead to a disruption of the NBD to WHD/HD2 interface by a ~90° rotation in the linker region between HD1 and WHD. The rotation can be seen by the last helix of HD1 (coloured bordeaux) relative to the first helix of WD1 (coloured olive). The pathogenic mutations may disrupt the integrity of the interface between NBD and WHD/HD2, preventing NLRP3 from entering the resting state of the autoinhibited conformation. The CINCA mutation Y861C is the only residue outside the NACHT domain with a validated pathogenic phenotype. As Y861 interacts with the acidic loop, its mutation could affect the dimer formation mediated by interface A.
Extended Data Fig. 9 Multiple sequence alignment of NLRP3 proteins.
Secondary structure elements are indicated for human NLRP3 as determined here. Subdomain boundaries are labelled and conserved sequence motifs are written italic. Residues in the decamer assembly interfaces A, B and C are labelled with asterisks. Smaller asterisks correspond to 10–50, larger to > 50 Å2 buried surface area, respectively. Pathogenic disease mutations FCAS1 (familial cold auto-inflammatory syndrome 1), CINCA (chronic infantile neurological cutaneous articular), MWS (Muckle-Wells syndrome) and CAPS (Cryopyrin associated periodic syndrome) are marked with circles as indicated. Sequences of human (UniProt accession number Q96P20), macaque (B0FPE9), mouse (Q8R4B8), rat (D4A523), and bovine (A6QLE5) NLRP3 proteins were aligned with MultAlin and the secondary structure annotated with ESPript50.
Extended Data Fig. 10 The CRID3-binding site in NLRP3.
a, CRID3 binds into a deep crevice that is spanned by subdomains NBD, HD1, WHD, HD2 and trLRR. Only the tertiary alcohol group reaches out of this binding cleft. b, Close-up of CRID binding to subdomains NBD, HD1 and WHD (left) and HD1, WHD, HD2 and trLRR (right). To visualize the binding sites, the other subdomains in each case were omitted. c, Density map around CRID3 displayed at 4 (black), 7 (red) and 10 (magenta) RMSD threshold. d, SPR measurements of NLRP3 mutants A228Q, R351T and R578A showed no binding to CRID3. e, Mutational analysis of the CRID3-binding interface in NLRP3 shows that all three mutants could not be activated by nigericin in cell-based assays. This suggests that the integrity of the binding site in between the NBD/HD1 and WHD/HD2 subdomains is critical for the activation mechanism of NLRP3. An ASC speck activation assay was used with HEK293T cells stably expressing an ASC-BFP fusion and transfected with a doxycycline-inducible NLRP3-T2A-mCherry construct containing either wild-type (wt) or CRID3-binding interface mutants. Data are mean ± SEM of n = 3 independent experiments (ns p > 0.05, * p < 0.05, **** p < 0.0001) (two-way ANOVA with Tukey’s multiple comparisons test).
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, gel source data for Extended Data Fig. 1b, gel source data for Extended Data Fig. 1h and gel source data for Extended Data Fig. 2d, e, f.
Supplementary Video 1
Central density. Video animation displaying the density in the central cavity of the NLRP3 decamer.
Supplementary Video 2
NLRP3 decamer. Video animation displaying a ribbon diagram of the NLRP3 monomer, its formation of an LRR intertwined dimer and the assembly of the decamer.
Rights and permissions
About this article
Cite this article
Hochheiser, I.V., Pilsl, M., Hagelueken, G. et al. Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604, 184–189 (2022). https://doi.org/10.1038/s41586-022-04467-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04467-w
This article is cited by
-
Structural basis of the subcortical maternal complex and its implications in reproductive disorders
Nature Structural & Molecular Biology (2024)
-
Mechanistic insights from inflammasome structures
Nature Reviews Immunology (2024)
-
Drugging the NLRP3 inflammasome: from signalling mechanisms to therapeutic targets
Nature Reviews Drug Discovery (2024)
-
Structural basis for the oligomerization-facilitated NLRP3 activation
Nature Communications (2024)
-
New Insights on NLRP3 Inflammasome: Mechanisms of Activation, Inhibition, and Epigenetic Regulation
Journal of Neuroimmune Pharmacology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.