Abstract
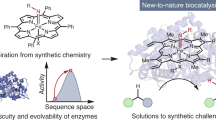
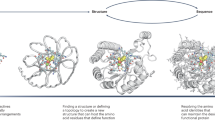
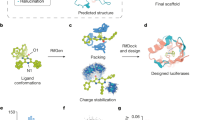
The ability to design efficient enzymes from scratch would have a profound effect on chemistry, biotechnology and medicine. Rapid progress in protein engineering over the past decade makes us optimistic that this ambition is within reach. The development of artificial enzymes containing metal cofactors and noncanonical organocatalytic groups shows how protein structure can be optimized to harness the reactivity of nonproteinogenic elements. In parallel, computational methods have been used to design protein catalysts for diverse reactions on the basis of fundamental principles of transition state stabilization. Although the activities of designed catalysts have been quite low, extensive laboratory evolution has been used to generate efficient enzymes. Structural analysis of these systems has revealed the high degree of precision that will be needed to design catalysts with greater activity. To this end, emerging protein design methods, including deep learning, hold particular promise for improving model accuracy. Here we take stock of key developments in the field and highlight new opportunities for innovation that should allow us to transition beyond the current state of the art and enable the robust design of biocatalysts to address societal needs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
Huffman, M. A. et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 366, 1255–1259 (2019). erratum 368, eabc1954 (2020).
Schober, M. et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat. Catal. 2, 909–915 (2019).
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Turner, N. J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5, 567–573 (2009).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Zeymer, C. & Hilvert, D. Directed evolution of protein catalysts. Annu. Rev. Biochem. 87, 131–157 (2018).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 57, 4143–4148 (2018).
Qu, G., Li, A., Acevedo-Rocha, C. G., Sun, Z. & Reetz, M. T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. Int. Ed. 59, 13204–13231 (2020).
Fernandez-Gacio, A., Uguen, M. & Fastrez, J. Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 21, 408–414 (2003).
Becker, S., Schmoldt, H. U., Adams, T. M., Wilhelm, S. & Kolmar, H. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts. Curr. Opin. Biotechnol. 15, 323–329 (2004).
Agresti, J. J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009 (2010).
Debon, A. et al. Ultrahigh-throughput screening enables efficient single-round oxidase remodelling. Nat. Catal. 2, 740–747 (2019).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Bryson, D. I. et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 13, 1253–1260 (2017).
Ravikumar, A., Arzumanyan, G. A., Obadi, M. K. A., Javanpour, A. A. & Liu, C. C. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell 175, 1946–1957 (2018).
Zhang, R. K. et al. Enzymatic assembly of carbon–carbon bonds via iron-catalysed sp3 C–H functionalization. Nature 565, 67–72 (2018).
Chen, K., Huang, X., Kan, S. B. J., Zhang, R. K. & Arnold, F. H. Enzymatic construction of highly strained carbocycles. Science 360, 71–75 (2018).
Biegasiewicz, K. F. et al. Photoexcitation of flavoenzymes enables a stereoselective radical cyclization. Science 364, 1166–1169 (2019).
Ji, P., Park, J., Gu, Y., Clark, D. S. & Hartwig, J. F. Abiotic reduction of ketones with silanes catalysed by carbonic anhydrase through an enzymatic zinc hydride. Nat. Chem. 13, 312–318 (2021).
Kiss, G., Celebi-Olcum, N., Moretti, R., Baker, D. & Houk, K. N. Computational enzyme design. Angew. Chem. Int. Ed. 52, 5700–5725 (2013).
Hilvert, D. Design of protein catalysts. Annu. Rev. Biochem. 82, 447–470 (2013).
Baker, D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 19, 1817–1819 (2010).
Jeschek, M. et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature 537, 661–665 (2016).
Zhao, J. et al. Genetic engineering of an artificial metalloenzyme for transfer hydrogenation of a self-immolative substrate in Escherichia coli’s periplasm. J. Am. Chem. Soc. 140, 13171–13175 (2018).
Rebelein, J. G. & Ward, T. R. In vivo catalyzed new-to-nature reactions. Curr. Opin. Biotechnol. 53, 106–114 (2018).
Hyster, T. K., Knorr, L., Ward, T. R. & Rovis, T. Biotinylated Rh(III) complexes in engineered streptavidin for accelerated asymmetric C–H activation. Science 338, 500–503 (2012). Demonstration that transition metal complexes embedded in protein hosts can work in synergy with amino acid side chains to accelerate a challenging C–H activation process.
Bhagi-Damodaran, A. et al. Why copper is preferred over iron for oxygen activation and reduction in haem-copper oxidases. Nat. Chem. 9, 257–263 (2017).
Yeung, N. et al. Rational design of a structural and functional nitric oxide reductase. Nature 462, 1079–1082 (2009).
Mirts, E. N., Petrik, I. D., Hosseinzadeh, P., Nilges, M. J. & Lu, Y. A designed heme-[4Fe–4S] metalloenzyme catalyzes sulfite reduction like the native enzyme. Science 361, 1098–1101 (2018). This study shows how the introduction of new functional elements into metalloproteins can generate artificial enzymes for challenging chemical tranformations that have thus far eluded synthetic catalysts.
Hill, R. B., Raleigh, D. P., Lombardi, A. & DeGrado, W. F. De novo design of helical bundles as models for understanding protein folding and function. Acc. Chem. Res. 33, 745–754 (2000).
Koder, R. L. & Dutton, P. L. Intelligent design: the de novo engineering of proteins with specified functions. Dalton Trans. 25, 3045–3051 (2006).
Faiella, M. et al. An artificial di-iron oxo-protein with phenol oxidase activity. Nat. Chem. Biol. 5, 882–884 (2009).
Smith, B. A. & Hecht, M. H. Novel proteins: from fold to function. Curr. Opin. Chem. Biol. 15, 421–426 (2011).
Zastrow, M. L., Peacock, A. F., Stuckey, J. A. & Pecoraro, V. L. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat. Chem. 4, 118–123 (2011).
Stenner, R., Steventon, J. W., Seddon, A. & Anderson, J. L. R. A de novo peroxidase is also a promiscuous yet stereoselective carbene transferase. Proc. Natl Acad. Sci. USA 117, 1419–1428 (2020).
Chino, M. et al. A de novo heterodimeric Due Ferri protein minimizes the release of reactive intermediates in dioxygen-dependent oxidation. Angew. Chem. Int. Ed. 56, 15580–15583 (2017).
Lombardi, A., Pirro, F., Maglio, O., Chino, M. & DeGrado, W. F. De novo design of four-helix bundle metalloproteins: one scaffold, diverse reactivities. Acc. Chem. Res. 52, 1148–1159 (2019).
Reig, J. A. et al. Alteration of the oxygen-dependent reactivity of de novo Due Ferri proteins. Nat. Chem. 4, 900–906 (2012). Demonstration that the catalytic function of de novo Due Ferri proteins can be altered through rational reprogramming of the metal coordination environment.
Salgado, E. N., Faraone-Mennella, J. & Tezcan, F. A. Controlling protein–protein interactions through metal coordination: assembly of a 16-helix bundle protein. J. Am. Chem. Soc. 129, 13374–13375 (2007).
Der, B. S. et al. Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer. J. Am. Chem. Soc. 134, 375–385 (2012).
Der, B. S., Edwards, D. R. & Kuhlman, B. Catalysis by a de novo zinc-mediated protein interface: implications for natural enzyme evolution and rational enzyme engineering. Biochemistry 51, 3933–3940 (2012).
Studer, S. et al. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 362, 1285–1288 (2018). This study uses a combination of design and evolution to transform a designed zinc-binding peptide into a globular metalloenzyme that accelerates ester hydrolysis with high efficiency.
Basler, S. et al. Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold. Nat. Chem. 13, 231–235 (2021). In this study, a de novo metalloenzyme is engineered to accelerate an abiological hetero-Diels–Alder reaction with high specificity and a catalytic proficiency that exceeds all previously characterized Diels–Alderases.
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Seyedsayamdost, M. R., Xie, J., Chan, C. T. Y., Schultz, P. G. & Stubbe, J. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: direct evidence for involvement of Y730 and Y731 in radical propagation. J. Am. Chem. Soc. 129, 15060–15071 (2007).
Faraldos, J. A. et al. Probing eudesmane cation−π interactions in catalysis by aristolochene synthase with non-canonical amino acids. J. Am. Chem. Soc. 133, 13906–13909 (2011).
Wu, Y. & Boxer, S. G. A critical test of the electrostatic contribution to catalysis with noncanonical amino acids in ketosteroid isomerase. J. Am. Chem. Soc. 138, 11890–11895 (2016).
Ortmayer, M. et al. Rewiring the ‘push–pull’ catalytic machinery of a heme enzyme using an expanded genetic code. ACS Catal. 10, 2735–2746 (2020).
Ortmayer, M. et al. A noncanonical tryptophan analogue reveals an active site hydrogen bond controlling ferryl reactivity in a heme peroxidase. JACS Au 1, 913–918 (2021).
Li, J. C., Liu, T., Wang, Y., Mehta, A. P. & Schultz, P. G. Enhancing protein stability with genetically encoded noncanonical amino acids. J. Am. Chem. Soc. 140, 15997–16000 (2018).
Green, A. P., Hayashi, T., Mittl, P. R. & Hilvert, D. A chemically programmed proximal ligand enhances the catalytic properties of a heme enzyme. J. Am. Chem. Soc. 138, 11344–11352 (2016).
Zhao, J., Burke, A. J. & Green, A. P. Enzymes with noncanonical amino acids. Curr. Opin. Chem. Biol. 55, 136–144 (2020).
Burke, A. J. et al. Design and evolution of an enzyme with a non-canonical organocatalytic mechanism. Nature 570, 219–223 (2019). Demonstrates how introducing noncanonical amino acids can expand the chemical reactivity and catalytic mechanisms accessible with designed enzymes.
Bolon, D. N. & Mayo, S. L. Enzyme-like proteins by computational design. Proc. Natl Acad. Sci. USA 98, 14274–14279 (2001).
Richter, F. et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J. Am. Chem. Soc. 134, 16197–16206 (2012).
Rajagopalan, S. et al. Design of activated serine-containing catalytic triads with atomic-level accuracy. Nat. Chem. Biol. 10, 386–391 (2014).
Moroz, Y. S. et al. New tricks for old proteins: single mutations in a nonenzymatic protein give rise to various enzymatic activities. J. Am. Chem. Soc. 137, 14905–14911 (2015).
Burton, A. J., Thomson, A. R., Dawson, W. M., Brady, R. L. & Woolfson, D. N. Installing hydrolytic activity into a completely de novo protein framework. Nat. Chem. 8, 837–844 (2016).
Drienovska, I., Mayer, C., Dulson, C. & Roelfes, G. A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue. Nat. Chem. 10, 946–952 (2018). Demonstrates that the introduction of noncanonical amino acids can open up new modes of reactivity within proteins.
Mayer, C., Dulson, C., Reddem, E., Thunnissen, A. W. H. & Roelfes, G. Directed evolution of a designer enzyme featuring an unnatural catalytic amino acid. Angew. Chem. Int. Ed. 58, 2083–2087 (2019).
Tramontano, A., Janda, K. D. & Lerner, R. A. Catalytic antibodies. Science 234, 1566–1570 (1986).
Wagner, J., Lerner, R. A. & Barbas, C. F. III. Efficient aldolase catalytic antibodies that use the enamine mechanism of natural enzymes. Science 270, 1797–1800 (1995).
Gouverneur, V. E. et al. Control of the exo and endo pathways of the Diels–Alder reaction by antibody catalysis. Science 262, 204–208 (1993).
Wentworth, P. Jr. et al. Antibody catalysis of the oxidation of water. Science 293, 1806–1811 (2001).
Hsieh, L. C., Yonkovich, S., Kochersperger, L. & Schultz, P. G. Controlling chemical reactivity with antibodies. Science 260, 337–339 (1993).
Hilvert, D. Critical analysis of antibody catalysis. Annu. Rev. Biochem. 69, 751–793 (2000).
Rothlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Privett, H. K. et al. Iterative approach to computational enzyme design. Proc. Natl Acad. Sci. USA 109, 3790–3795 (2012).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010). Computational design and experimental characterization of enzymes catalysing a bimolecular Diels–Alder reaction, an important carbon–carbon bond-forming process.
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013). Demonstrates that artificial enzymes can be evolved to accelerate elementary chemical reactions with efficiencies comparable to natural enzymes.
Preiswerk, N. et al. Impact of scaffold rigidity on the design and evolution of an artificial Diels–Alderase. Proc. Natl Acad. Sci. USA 111, 8013–8018 (2014).
Obexer, R. et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem. 9, 50–56 (2017). Ultrahigh-throughput screening facilitates the development of artificial enzymes with efficiencies comparable to natural systems.
Crawshaw, R. et al. Engineering an efficient and enantioselective enzyme for the Morita–Baylis–Hillman reaction. Nat. Chem. 14, 313–320 (2022). A demonstration that laboratory evolution of designed enzymes can deliver sophisticated active sites to accelerate complex nonbiological transformations.
Otten, R. et al. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 370, 1442–1446 (2020).
Broom, A. et al. Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico. Nat. Commun. 11, 4808 (2020).
Althoff, E. A. et al. Robust design and optimization of retroaldol enzymes. Protein Sci. 21, 717–726 (2012).
Giger, L. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol. 9, 494–498 (2013).
Eiben, C. B. et al. Increased Diels–Alderase activity through backbone remodeling guided by Foldit players. Nat. Biotechnol. 30, 190–192 (2012).
Bjelic, S. et al. Computational design of enone-binding proteins with catalytic activity for the Morita–Baylis–Hillman reaction. ACS Chem. Biol. 8, 749–757 (2013).
Kiss, G., Rothlisberger, D., Baker, D. & Houk, K. N. Evaluation and ranking of enzyme designs. Protein Sci. 19, 1760–1773 (2010).
Frushicheva, M. P., Cao, J., Chu, Z. T. & Warshel, A. Exploring challenges in rational enzyme design by simulating the catalysis in artificial Kemp eliminase. Proc. Natl Acad. Sci. USA 107, 16869–16874 (2010).
Bunzel, H. A. et al. Evolution of dynamical networks enhances catalysis in a designer enzyme. Nat. Chem. 13, 1017–1022 (2021).
Weitzner, B. D., Kipnis, Y., Daniel, A. G., Hilvert, D. & Baker, D. A computational method for design of connected catalytic networks in proteins. Protein Sci. 28, 2036–2041 (2019).
Davey, J. A., Damry, A. M., Goto, N. K. & Chica, R. A. Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 13, 1280–1285 (2017).
Pan, X. et al. Expanding the space of protein geometries by computational design of de novo fold families. Science 369, 1132–1136 (2021).
Huang, P. S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320–327 (2016).
Dou, J. et al. De novo design of a fluorescence-activating β-barrel. Nature 561, 485–491 (2018).
Wei, K. Y. et al. Computational design of closely related proteins that adopt two well-defined but structurally divergent folds. Proc. Natl Acad. Sci. USA 117, 7208–7215 (2020).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). Development of AlphaFold, a deep learning algorithm for accurate prediction of protein structure from primary sequence.
Hiranuma, N. et al. Improved protein structure refinement guided by deep learning based accuracy estimation. Nat. Commun. 12, 1340 (2021).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). Development of RoseTTAFold, a freely available deep learning programme for fast and accurate prediction of protein structure.
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Anishchenko, I. et al. De novo protein design by deep network hallucination. Nature 600, 547–552 (2021).
Mazurenko, S., Prokop, Z. & Damborsky, J. Machine learning in enzyme engineering. ACS Catal. 10, 1210–1223 (2020).
Ma, E. J. et al. Machine-directed evolution of an imine reductase for activity and stereoselectivity. ACS Catal. 11, 12433–12445 (2021).
Bedbrook, C. N., Yang, K. K., Rice, A. J., Gradinaru, V. & Arnold, F. A. Machine learning to design integral membrane channelrhodopsins for efficient eukaryotic expression and plasma membrane localization. PLoS Comput. Biol. 13, e1005786 (2017).
Wu, Z., Kan, S. B. J., Lewis, R. D., Wittmann, B. J. & Arnold, F. H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl Acad. Sci. USA 116, 8852–8858 (2019).
Tischer, D. et al. Design of proteins presenting discontinuous functional sites using deep learning. Preprint at bioRxiv https://doi.org/10.1101/2020.11.29.402743.
Russ, W. P. et al. An evolution-based model for designing chorismate mutase enzymes. Science 369, 440–445 (2020).
Wang, J. et al. Deep learning methods for designing proteins scaffolding functional sites. Preprint at bioRxiv https://doi.org/10.1101/2021.11.10.468128.
Hayashi, T. et al. Capture and characterization of a reactive haem–carbenoid complex in an artificial metalloenzyme. Nat. Catal. 1, 578–584 (2018).
Carminati, D. M. & Fasan, R. Stereoselective cyclopropanation of electron-deficient olefins with a cofactor redesigned carbene transferase featuring radical reactivity. ACS Catal. 9, 9683–9687 (2019).
Erkkila, A., Majander, I. & Pihko, P. M. Iminium catalysis. Chem. Rev. 107, 5416–5470 (2007).
Mukherjee, S., Yang, J. W., Hoffmann, S. & List, B. Asymmetric enamine catalysis. Chem. Rev. 107, 5471–5569 (2007).
Doyle, A. G. & Jacobsen, E. N. Small-molecule H-bond donors in asymmetric catalysis. Chem. Rev. 107, 5713–5743 (2007).
Wurz, R. P. Chiral dialkylaminopyridine catalysts in asymmetric synthesis. Chem. Rev. 107, 5570–5595 (2007).
Beeson, T. D., Mastracchio, A., Hong, J. B., Ashton, K. & Macmillan, D. W. Enantioselective organocatalysis using SOMO activation. Science 316, 582–585 (2007).
St-Jacques, A. D., Eyahpaise, M.-È. C. & Chica, R. A. Computational design of multisubstrate enzyme specificity. ACS Catal. 9, 5480–5485 (2019).
Davey, J. A. & Chica, R. A. Multistate approaches in computational protein design. Protein Sci. 21, 1241–1252 (2012).
Acknowledgements
We thank the European Research Council (ERC Starter Grant, no. 757991 to A.P.G.), the Biotechnology and Biological Sciences Research Council (David Phillips Fellowship BB/M027023/1 to A.P.G.), UK Research and Innovation (Future Leader Fellowship MR/T041722/1 to S.L.L.), the Swiss National Science Foundation (D.H.), ETH Zürich (D.H.) and the Howard Hughes Medical Institute (D.B.) for generous support.
Author information
Authors and Affiliations
Contributions
All authors discussed the content of the manuscript, including selection of key studies highlighted and opportunities for future innovations. R.C., S.B. and C.L. prepared the figures. S.L.L., D.B., D.H. and A.P.G. wrote the manuscript text.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lovelock, S.L., Crawshaw, R., Basler, S. et al. The road to fully programmable protein catalysis. Nature 606, 49–58 (2022). https://doi.org/10.1038/s41586-022-04456-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04456-z
This article is cited by
-
Enzyme engineering for functional lipids synthesis: recent advance and perspective
Bioresources and Bioprocessing (2024)
-
An artificial protein modulator reprogramming neuronal protein functions
Nature Communications (2024)
-
Engineered enzymes for the synthesis of pharmaceuticals and other high-value products
Nature Synthesis (2024)
-
A non-canonical nucleophile unlocks a new mechanistic pathway in a designed enzyme
Nature Communications (2024)
-
Designer catalytic nanopores meet PET nanoparticles
Nature Catalysis (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.