Abstract
Disruption of susceptibility (S) genes in crops is an attractive breeding strategy for conferring disease resistance1,2. However, S genes are implicated in many essential biological functions and deletion of these genes typically results in undesired pleiotropic effects1. Loss-of-function mutations in one such S gene, Mildew resistance locus O (MLO), confers durable and broad-spectrum resistance to powdery mildew in various plant species2,3. However, mlo-associated resistance is also accompanied by growth penalties and yield losses3,4, thereby limiting its widespread use in agriculture. Here we describe Tamlo-R32, a mutant with a 304-kilobase pair targeted deletion in the MLO-B1 locus of wheat that retains crop growth and yields while conferring robust powdery mildew resistance. We show that this deletion results in an altered local chromatin landscape, leading to the ectopic activation of Tonoplast monosaccharide transporter 3 (TaTMT3B), and that this activation alleviates growth and yield penalties associated with MLO disruption. Notably, the function of TMT3 is conserved in other plant species such as Arabidopsis thaliana. Moreover, precision genome editing facilitates the rapid introduction of this mlo resistance allele (Tamlo-R32) into elite wheat varieties. This work demonstrates the ability to stack genetic changes to rescue growth defects caused by recessive alleles, which is critical for developing high-yielding crop varieties with robust and durable disease resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The sequencing data obtained in this study have been deposited in the Genome Sequence Archive (GSA) database in the BIG Data Center (https://ngdc.cncb.ac.cn/) under accession number PRJCA005687. Chinese Spring wheat reference genome RefSeq v1.1 is available on IWGSC (http://www.wheatgenome.org/). Transcriptome data from different wheat tissues are from ref. 29. Source data are provided with this paper.
References
van Schie, C. C. & Takken, F. L. Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581 (2014).
Schulze-Lefert, P. & Vogel, J. Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348 (2000).
Büschges, R. et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705 (1997).
Consonni, C. et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720 (2006).
van Esse, H. P., Reuber, T. L. & van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 225, 70–86 (2020).
Li, W., Deng, Y., Ning, Y., He, Z. & Wang, G.-L. Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603 (2020).
Dangl, J. L., Horvath, D. M. & Staskawicz, B. J. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013).
Deng, Y. et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965 (2017).
Saintenac, C. et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50, 368–374 (2018).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
Oliva, R. et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350 (2019).
Lapin, D. & Van den Ackerveken, G. Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 18, 546–554 (2013).
Vogel, J. P., Raab, T. K., Schiff, C. & Somerville, S. C. PMR6, a pectate lyase–like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106 (2002).
Eckardt, N. A. Plant disease susceptibility genes? Plant Cell 14, 1983–1986 (2002).
Kim, M. C. et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451 (2002).
Devoto, A. et al. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34993–35004 (1999).
Kusch, S. & Panstruga, R. mlo-based resistance: an apparently universal "weapon" to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189 (2017).
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Bai, Y. et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol. Plant Microbe Interact. 21, 30–39 (2008).
Humphry, M., Consonni, C. & Panstruga, R. mlo-based powdery mildew immunity: silver bullet or simply non-host resistance? Mol. Plant Pathol. 7, 605–610 (2006).
Piffanelli, P. et al. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891 (2004).
Acevedo-Garcia, J., Kusch, S. & Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204, 273–281 (2014).
Appiano, M. et al. Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol. 15, 257 (2015).
Singh, R. P. et al. Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322 (2016).
Acevedo-Garcia, J. et al. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 15, 367–378 (2017).
Bogdanove, A. J. & Voytas, D. F. TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846 (2011).
Ramírez-González, R. H. et al. The transcriptional landscape of polyploid wheat. Science 361, eaar6089 (2018).
Wormit, A. et al. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18, 3476–3490 (2006).
Bieluszewski, T., Xiao, J., Yang, Y. & Wagner, D. PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 21, S1360–S1385 (2021).
Kajimura, T., Mizuno, N. & Takumi, S. Utility of leaf senescence-associated gene homologs as developmental markers in common wheat. Plant Physiol. Biochem. 48, 851–859 (2010).
Consonni, C. et al. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 152, 1544–1561 (2010).
Gao, C. Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635 (2021).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun.7, 12617–12624 (2016).
Liang, Z. et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun.8, 14261–14265 (2017).
Jansen, M., Jarosch, B. & Schaffrath, U. The barley mutant emr1 exhibits restored resistance against Magnaporthe oryzae in the hypersusceptible mlo-genetic background. Planta 225, 1381–1391 (2007).
Li, S. et al. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28, 2866–2883 (2016).
Shan, Q. et al. Targeted genome modification of crop plants using a CRISPR–Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Liang, Z. et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430 (2018).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Wang, Z. et al. Genetic and physical mapping of powdery mildew resistance gene MlHLT in Chinese wheat landrace Hulutou. Theor. Appl. Genet. 128, 365–373 (2015).
Shen, Q. H. et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103 (2007).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 (2013).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Weber, B., Jamge, S. & Stam, M. 3C in maize and Arabidopsis. Methods Mol. Biol. 1675, 247–270 (2018).
Krijger, P. H., Geeven, G., Bianchi, V., Hilvering, C. R., & de Laat, W. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods 170, 17–32. (2020).
Ricci, W. A. et al. Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5, 1237–1249 (2019).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Kim, D. & Sung, S. Vernalization-triggered intragenic chromatin-loop formation by long noncoding RNAs. Dev. Cell 40, 302–312 (2017).
Paolacci, A. R., Tanzarella, O. A., Porceddu, E. & Ciaffi, M. Identification and validation of reference genes for quantitative RT–PCR normalization in wheat. BMC Mol. Biol. 10, 11 (2009).
Bajic, M., Maher, K. A. & Deal, R. B. Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol. Biol. 1675, 183–201 (2018).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Xiao, J. et al. Cis- and trans-determinants of epigenetic silencing by Polycomb Repressive Complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552 (2017).
Gou, J. Y. et al. Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27, 1755–1770 (2015).
Qin, D. et al. Characterization and fine mapping of a novel barley Stage Green-Revertible Albino gene (HvSGRA) by bulked segregant analysis based on ssr assay and specific length amplified fragment sequencing. BMC Genomics 16, 838–851 (2015).
Ni, Z. et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331 (2009).
Acknowledgements
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020101 to J.-L.Q., XDA24020102 to C.G., XDA24010204 to J.X., XDA24020310 to Y.W., XDPB16 to J.-L.Q.), the National Natural Science Foundation of China (31788103 to C.G., 32001891 to S.L., 31970529 to J.X. and 31971370 to K.C.) and the Key Research Program of Frontier Sciences (QYZDY-SSW-SMC030 to C.G.) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2020000003 to Y.W.).
Author information
Authors and Affiliations
Contributions
C.G. and J.-L.Q. conceived and conceptualized the study. C.G. and J.-L.Q. designed the experiments. S.L., D.L. and Y.Z. performed most of the experiments. S.L., Y.W. and J.X. prepared the figures. S.L. and Y.Z. performed the powdery mildew infection experiment. D.L., B. Li, Y. Lei, J.L. and K.C carried out genome-editing experiments and mutant identification. M.D. conducted the 4C and 3C experiments. L.Z. and J.X. conducted the CUT&Tag, ATAC-seq and bioinformatics analyses. B. Lv, and Y. Liang performed the marker-assisted selection (MAS) and powdery mildew microscopic analyses. S.L. and Y.W. characterized the phenotypes of mutant plants. Y.C. and Z.L. carried out traditional breeding and field trials. J.-L.Q., C.G., S.L. and J.X wrote the manuscript. All authors commented on the results and contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.G., J.-L.Q., S.L. and Y.W. have filed patent applications based on the work published here.
Peer review
Peer review information
Nature thanks Wendy Harwood, Nian Wang and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Genotyping by agarose gel electrophoresis and Sanger sequencing.
a, Detection of the transgene in the Tamlo-R32 mutants by PCR using five independent primer sets. Plasmid DNA of the TALEN vector was used as a positive control. b, Schematic diagram of the structure of the wheat TaMLO1 gene. Green rectangles and solid black lines represent exons and introns, respectively. The conserved TALEN target site within TaMLO1 is indicated by the red vertical line. Black arrows denote the positions and orientations of the three pairs of gene-specific primers (F1/R1, F2/R2, F3/R3) for amplifying TaMLO-A1, TaMLO-B1 and TaMLO-D1, respectively. c, Agarose gel electrophoresis of TaMLO1 amplicons from genomic DNA of the BW wild-type (upper) and Tamlo-R32 mutant (lower) using gene-specific primers. d, Agarose gel electrophoresis of the PCR products amplified by the primer pairs F2/R2 (upper) and F4/R4 (lower) from genomic DNA of the wild type and Tamlo-R32 mutants. The positions and orientations of the F4/R4 primer pairs are denoted by black arrows in Fig. 1g. e, DNA sequence of the edited sites in Tamlo-R32. Blue letters indicate inserted sequences. Red letters indicate the original sequence. Black vertical lines indicate the target site. The black dotted line indicates the deleted region. For a, c, d, experiments were repeated 3 times with the same results.
Extended Data Fig. 2 The Tamlo-R32 mutant displays no yield penalties when grown in the field.
a–d, The agronomic traits, including grain yield per plant (a), thousand kernel weight (b), tiller number per plant (c) and grain number per spike (d) were evaluated in field conditions in two wheat-growing areas in the North China Plain, Beijing and Zhaoxian in Hebei Province in 2019 and 2020. For box plots, the box limits indicate the twenty-fifth and seventy-fifth percentiles, the whiskers indicate the full range of the data, and the center line indicates the median. Individual data points are plotted. n represents the sample size. Statistical significance was determined by two-tailed Mann-Whitney tests or two-tailed Student’s t-tests. P values are indicated
Extended Data Fig. 3 Macroscopic powdery mildew infection phenotypes of F2 plants from various crosses.
a, b, Representative detached leaves of the F2 generations of Tamlo-R32×Tamlo-aaBBdd (a) and Tamlo-R32×Tamlo-aabbdd (b) are shown seven days after inoculation of Bgt isolate E09. Red triangles indicate resistant plants. Scale bar, 1 cm.
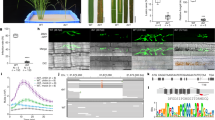
Extended Data Fig. 4 Expression patterns and chromatin landscapes of genes around the large deletion.
a, Expression of TaTMT3B and 11 other nearby genes in different tissues; data are from a previous publication30. Genes within the deleted region are indicated. b, c, Amino acid sequence alignment between homeologs on the A, B and D genomes of downregulated genes in the deletion region. d, Expression levels of TaTMT3B in different tissues of Tamlo-R32 and WT plants measured by quantitative RT-PCR. Results are normalized to TaPAGE gene. n.d., not detected. Data are means ± s.d., of three independent RNA preparations from biological replicates. e, Chromatin accessibility, and histone modification profiles in the TaTMT3B-MLO-B1 region in leaf tissue of Chinese Spring wheat. The Integrative Genomics Viewer (IGV) views show the various chromatin status profiles near TaTMT3B. The y-axis represents signal enrichment computed from reads at each position normalized to the total number of reads (RPKM). The dark shading indicates regions with either repressive (H3K27me3) or active (H3K4me3, H3K36me3, H3K27ac) histone modifications (a-f). f, Schematic illustration of a possible model for regulation of the activation of TaTMT3B expression in Tamlo-R32
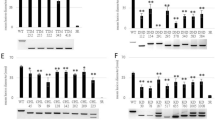
Extended Data Fig. 5 Expression levels of twenty genes around the large deletion measured by quantitative RT-PCR.
a, Schematic diagram of the gene distribution around the large B-genome deletion identified in Tamlo-R32. b, Expression levels of twenty genes of the B genome were measured by quantitative RT-PCR in both wild-type Bobwhite and Tamlo-R32 mutant leaves. Results are normalized to TaPARG gene and the expression level of gene in wild-type BW plants was set at one except TaTMT3B. n.d., not detected. Data are means ± s.d. of three independent RNA preparations from biological replicates
Extended Data Fig. 6 Wheat and Arabidopsis mlo mutants overexpressing TMT3 maintain powdery mildew resistance.
a, Targeted knockout of TaTMT3B by CRISPR-Cas9 in the Tamlo-R32 background. Blue letters indicate TaTMT3B sgRNA. The PAM sequence is highlighted in red. The numbers on the right show the type of mutation and how many nucleotides are involved, with “-” indicating deletion of the given number of nucleotides. b, Expression levels of TaTMT3 in TaTMT3B-overexpressors in the Tamlo-aabbdd mutant background, assessed by quantitative RT-PCR. The primers were designed to detect complete TaTMT3 transcripts. The results are normalized to TaACTIN, and expression of the gene in KN199 (wild type) is set at one. Data are means ± s.d., of three independent RNA preparations from biological replicates. c, Macroscopic infection phenotypes of representative detached leaves of the indicated wheat plants seven days after inoculation with Bgt isolate E09. Scale bar, 1 cm. d, Micrographs of microcolony formation by Bgt on wheat leaves of the indicated genotypes three days postinoculation. Powdery mildew spores and colonies were stained with Coomassie blue. Scale bars, 100 μm. e, Percentages of microcolonies formed from the total number of germinated spores of Bgt on the leaves of the indicated wheat plants. f, Expression levels of AtTMT3 in TMT3-overexpressors in the Atmlo2/6/12 background measured by quantitative RT-PCR. The primers were designed to detect both transgenic and endogenous AtTMT3 transcripts. The results are normalized to Arabidopsis AtACTIN8, and the expression level of the gene in WT was set at one. Data are means ± s.d. of three independent RNA preparations from biological replicates. g, Detached rosette leaves of the indicated 7-week-old Arabidopsis plants grown under long-day condition were laid out. Scale bar, 1 cm. h, Chlorophyll content of 6th rosette leaves of 7-week-old Arabidopsis plants grown under long-day conditions. The Tamlo-aabbdd mutants in b–e is in the KN199 background. Data are means of three biological replicates. Error bars represent means ± s.d. P values are indicated. i, Macroscopic infection phenotypes of representative detached leaves of the indicated Arabidopsis plants seven days after inoculation with G. orontii Scale bar, 1 cm. j, Micrographs of microcolony formation by G. orontii on Arabidopsis leaves of the indicated genotypes three days post-inoculation. Powdery mildew spores and colonies were stained with Coomassie blue. Scale bars, 100 μm. k, Percentages of microcolonies formed from the total number of germinated spores of G. orontii on leaves of indicated Arabidopsis plants. More than 1000 germinated spores per genotype per experiment were examined 72 h after inoculation in e and k. Data are means of three independent experiments. Error bars represent means ± s.d. Statistical significance in e, h, k was determined by two-tailed Mann-Whitney tests or two-tailed Student’s t-tests
Extended Data Fig. 7 Detection of transgene-free mutants.
Outcome of tests for transgene-free mutants using five primer sets in 31 mutant plants in Extended Data Table 2. Lanes labelled WT and plasmid show the PCR fragments amplified from a WT plant and plasmid constructs pJIT163-Ubi-Cas9 and pU6-gRNA vector respectively. KN-WT, XN-WT, XY-WT and S-WT indicate elite wheat varieties KN199, XN511, XY60 and S4185. Experiments were repeated 3 times with the same results.
Supplementary information
Supplementary Information
This file contains the uncropped images of agarose gels (Supplementary Fig. 1) and a list of primers used in the study (Supplementary Table 1).
Rights and permissions
About this article
Cite this article
Li, S., Lin, D., Zhang, Y. et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460 (2022). https://doi.org/10.1038/s41586-022-04395-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04395-9
This article is cited by
-
Potato calcium sensor modules StCBL3-StCIPK7 and StCBL3-StCIPK24 negatively regulate plant immunity
BMC Plant Biology (2024)
-
Impact of biotic stresses on the Brassicaceae family and opportunities for crop improvement by exploiting genotyping traits
Planta (2024)
-
CRISPR/Cas-mediated germplasm improvement and new strategies for crop protection
Crop Health (2024)
-
CRISPR/Cas genome editing in plants: mechanisms, applications, and overcoming bottlenecks
Functional & Integrative Genomics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.