Abstract
Mutations in the protein kinase PINK1 lead to defects in mitophagy and cause autosomal recessive early onset Parkinson’s disease1,2. PINK1 has many unique features that enable it to phosphorylate ubiquitin and the ubiquitin-like domain of Parkin3,4,5,6,7,8,9. Structural analysis of PINK1 from diverse insect species10,11,12 with and without ubiquitin provided snapshots of distinct structural states yet did not explain how PINK1 is activated. Here we elucidate the activation mechanism of PINK1 using crystallography and cryo-electron microscopy (cryo-EM). A crystal structure of unphosphorylated Pediculus humanus corporis (Ph; human body louse) PINK1 resolves an N-terminal helix, revealing the orientation of unphosphorylated yet active PINK1 on the mitochondria. We further provide a cryo-EM structure of a symmetric PhPINK1 dimer trapped during the process of trans-autophosphorylation, as well as a cryo-EM structure of phosphorylated PhPINK1 undergoing a conformational change to an active ubiquitin kinase state. Structures and phosphorylation studies further identify a role for regulatory PINK1 oxidation. Together, our research delineates the complete activation mechanism of PINK1, illuminates how PINK1 interacts with the mitochondrial outer membrane and reveals how PINK1 activity may be modulated by mitochondrial reactive oxygen species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The coordinates and crystallographic structure factors for PhPINK1(D334A) have been deposited at the PDB under accession code 7T3X, and EM models and maps under accession codes 7T4M (PhPINK1(D357A) dodecamer; Electron Microscopy Data Bank (EMDB): EMD-25680), 7T4N (PhPINK1(D357A) dimer; EMDB: EMD-25681), 7T4L (WT phosphorylated PhPINK1 dimer with extended αC helix in chain B; EMDB: EMD-25679) and 7T4K (WT phosphorylated PhPINK1 dimer with kinked αC helix in chain B; EMDB: EMD-25678). We also deposited the map for WT phosphorylated PhPINK1 dimer before 3D variability analysis (EMDB: EMD-25677). Uncropped versions of all gels and blots are provided in Supplementary Fig. 1. Source data are provided with this paper.
Code availability
The MATLAB script that was used to perform the Kolmogorov–Smirnov test to analyse lattice light-sheet imaging data is provided in the Supplementary Data.
Change history
15 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-04591-7
References
Valente, E. M. et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Pickrell, A. M. & Youle, R. J. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015).
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating serine 65. Open Biol. 2, 120080 (2012).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014).
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015).
Gladkova, C. et al. An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J. 36, 3555–3572 (2017).
Schubert, A. F. et al. Structure of PINK1 in complex with its substrate ubiquitin. Nature 552, 51–56 (2017).
Kumar, A. et al. Structure of PINK1 and mechanisms of Parkinson’s disease-associated mutations. eLife 6, e29985 (2017).
Okatsu, K. et al. Structural insights into ubiquitin phosphorylation by PINK1. Sci. Rep. 8, 10382 (2018).
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression and mortality. Neurology 17, 427–442 (1967).
Corti, O., Lesage, S. & Brice, A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol. Rev. 91, 1161–1218 (2011).
Nguyen, T. N., Padman, B. S. & Lazarou, M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 26, 733–744 (2016).
Harper, J. W., Ordureau, A. & Heo, J.-M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108 (2018).
Lazarou, M., Jin, S. M., Kane, L. A. & Youle, R. J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 (2012).
Okatsu, K. et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288, 36372–36384 (2013).
Wauer, T., Simicek, M., Schubert, A. F. & Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374 (2015).
Kumar, A. et al. Parkin–phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat. Struct. Mol. Biol. 24, 475–483 (2017).
Gladkova, C., Maslen, S. L., Skehel, J. M. & Komander, D. Mechanism of parkin activation by PINK1. Nature 559, 410–414 (2018).
Sauvé, V. et al. Mechanism of parkin activation by phosphorylation. Nat. Struct. Mol. Biol. 25, 623–630 (2018).
Woodroof, H. I. et al. Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 1, 110012 (2011).
Taylor, S. S. & Kornev, A. P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci 36, 65–77 (2011).
Patel, O. et al. Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association. Nat. Commun. 8, 1157 (2017).
Ha, B. H. & Boggon, T. J. The crystal structure of pseudokinase PEAK1 (Sugen kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization. J. Biol. Chem. 293, 1642–1650 (2018).
Sekine, S. et al. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol. Cell 73, 1028–1043 (2019).
Hasson, S. A. et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 (2013).
Rasool, S. et al. Mechanism of PINK1 activation by autophosphorylation and insights into assembly on the TOM complex. Mol. Cell 82, 44–59 (2022).
Rasool, S. et al. PINK1 autophosphorylation is required for ubiquitin recognition. EMBO Rep. 19, e44981 (2018).
Aerts, L., Craessaerts, K., Strooper, B. D. & Morais, V. A. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J. Biol. Chem. 290, 2798–2811 (2015).
Aerts, L., Craessaerts, K., Strooper, B. D. & Morais, V. A. In vitro comparison of the activity requirements and substrate specificity of human and triboleum castaneum PINK1 orthologues. PLoS ONE 11, e0146083 (2016).
Okatsu, K. et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3, 1016 (2012).
Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Soylu, İ. & Marino, S. M. Cy‐preds: an algorithm and a web service for the analysis and prediction of cysteine reactivity. Proteins Struct. Funct. Bioinform. 84, 278–291 (2016).
Shrestha, S. et al. A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily. Sci. Signal. 13, eaax6313 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Tunyasuvunakool, K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021).
Ovchinnikov, S., Mirdita, M. & Steinegger, M. ColabFold—making protein folding accessible to all via Google Colab (v1.0-alpha). Zenodo https://doi.org/10.5281/zenodo.5123297 (2021).
Pollock, L., Jardine, J., Urbé, S. & Clague, M. J. The PINK1 repertoire: not just a one trick pony. Bioessays 43, 2100168 (2021).
Hertz, N. T. et al. A neo-substrate that amplifies catalytic activity of Parkinson’s-disease-related kinase PINK1. Cell 154, 737–747 (2013).
Lowe, E. D. et al. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16, 6646–6658 (1997).
Berrow, N. S. et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45 (2007).
Aragão, D. et al. MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 25, 885–891 (2018).
McPhillips, T. M. et al. Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Yang, Z. et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 179, 269–278 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Slater, M. et al. Achieve the protein expression level you need with the mammalian HaloTag 7 flexi vectors. Promega Notes 100, 16–18 (2008).
Yamano, K., Fogel, A. I., Wang, C., Bliek, A. M. V. D. & Youle, R. J. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 3, e01612 (2014).
McArthur, K. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359, eaao6047 (2018).
Wang, W. et al. Atomic structure of human TOM core complex. Cell Discov. 6, 67 (2020).
Acknowledgements
We thank the members of the Ubiquitin Signalling Division at WEHI and I. Lucet (WEHI) and M. Lazarou (Monash) for reagents, advice and comments on the manuscript; J. Newman (CSIRO C3 Crystallisation Centre) for help with crystallization; and the staff at the WEHI Information Technology Services and the WEHI Research Computing Platform for providing facilities and support. This work was funded by WEHI, an NHMRC Investigator Grant (GNT1178122 to D.K.), the Michael J. Fox Foundation and Shake-It-Up Australia (to D.K. and G.D.). A.G. is a CSL Centenary Fellow. G.D. is supported by a fellowship from the Bodhi Education Fund and NHMRC Ideas grant (GNT2004446). Z.Y.G. is supported by an Australian Government Research Training Program Scholarship. Research was further supported by an NHMRC Independent Research Institutes Infrastructure Support Scheme grant (361646) and Victorian State Government Operational Infrastructure Support grant. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. We also acknowledge use of the Titan Krios and other facilities at the Ian Holmes Imaging Centre at the Bio21 Molecular Science and Biotechnology Institute.
Author information
Authors and Affiliations
Contributions
D.K. conceived the project. Z.Y.G. designed and performed all experiments. S.C. and G.D. contributed to the design of cell biology experiments. D.K. and T.R.C. contributed to crystallography. S.A.C. performed mass spectrometry experiments. T.R.C. performed SEC–MALS experiments. S.C., M.J.M., N.D.G. and K.L.R. performed and analysed lattice light sheet microscopy experiments. A.F.S. contributed ideas, advice and preliminary data. A.L. and A.G. performed EM data collection, A.G. performed data processing and map calculations and Z.Y.G. performed model building. D.K. and Z.Y.G. analysed the data and wrote the manuscript with contributions from all of the authors.
Corresponding author
Ethics declarations
Competing interests
D.K. serves on the Scientific Advisory Board of BioTheryX Inc. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Tony Hunter, Titia Sixma and Richard Youle for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Crystal structures of unphosphorylated PINK1.
a, Constructs used in crystal structures of TcPINK1 (PDB: 5OAT11 and 5YJ912). Phosphomimetics substituted for Ser205 (equivalent to Ser202 in PhPINK1) and other residues, although one structure (PDB: 5OAT) still showed phosphorylation at Ser207 (equivalent to Ser204 in PhPINK1, and Ser230 in HsPINK1, see below). Constructs were further engineered as indicated and described11,12. b, Depiction of TcPINK1 crystal structures, with key features indicated. c, 2|Fo|–|Fc| electron density contoured at 1.5σ, covering the PhPINK1(D334A) molecule in the asymmetric unit. Right, detail for regions of interest including the αC helix and Ser202-containing loop and the N-helix. d, Our crystal structure of unphosphorylated PhPINK1(D334A) (compare Fig. 1) in the same orientation as in b, for comparison. e, Two different zoomed views of the ATP-binding site of unphosphorylated PhPINK1, with an ATP molecule modelled in from PDB 2PHK (ref. 42) as before10. Left, highlighting key features common to most active kinases, including the P-loop (grey), αC helix (orange) and activation segment (pink). The DFG and HRD motifs (dotted outlines) which include Asp357 involved in coordination of Mg2+, and the substrate-binding Asp334 involved in phosphoryl transfer (and mutated to Ala in the crystal structure) are indicated. Glu214 and Lys193 form a crucial salt bridge to coordinate ATP. Right, assembled catalytic (C) and regulatory (R) spines24 in red and blue, respectively, indicative of an active kinase conformation. f, Crystal contact involving N-helix residue Trp129, which binds to the N-lobe of a symmetry related molecule. ASU, asymmetric unit.
Extended Data Fig. 2 The N-helix and its role in keeping PINK1 monomeric.
a, The pseudokinases SgK223 (PDB: 5VE6) and SgK269 (PDB: 6BHC) were structurally characterised only recently25,26, and contain a CTR similar to PINK1 that is complemented by an N-helix, in a highly analogous fashion now shown for PINK1. While conceptually similar, the organization of the N-helix on the CTR is however distinct. SgK223 and SgK269 utilize a cross-architecture to provide a dimerization interface, whereas the orientation in PINK1 is parallel to the αK helix of the CTR. b, SEC–MALS analysis of PhPINK1 with (amino acids 115–575) or without (amino acids 143–575) the N-helix. The shorter PhPINK1 construct tends to form less well-defined dimers, whereas the longer variant is a monomer. Experiments were performed three times with identical results. c, Previous TcPINK1 and PhPINK1 structures dimerized in the crystal lattice through the CTR, reflecting an available interaction surface since the N-helix was missing. TcPINK1 structures dimerized identically (left, only shown for PDB 5OAT11). The relative orientation of PhPINK1 molecules in PDB 6EQI10 was different to TcPINK1 (middle). Right, PhPINK1(D334A) structure including the N-helix, for comparison. d, Schematic of the N-helix–CTR interaction, and situation in previous PINK1 structures, as identified in structures and in SEC–MALS.
Extended Data Fig. 3 PhPINK1(D357A) oligomerization enables EM studies.
a, Elution profile of the PhPINK1(D357A) mutant during purification by SEC. Data shown is from a representative experiment of three runs. b, Thermal denaturation studies of purified PhPINK1 (amino acids 115–575) variants. The crystallized D334A mutant has a melting curve profile and melting temperature similar to WT PhPINK1. PhPINK1(D357A) shows an unusual profile with a high secondary melting temperature. Technical duplicates were measured in three independent experiments. Average melting temperatures are indicated. c, Negative stain EM analysis of PhPINK1(D357A) revealed a highly ordered oligomer suitable for cryo-EM studies. Minimal processing was performed in RELION (v.3.1)51, and a subset of the resulting 2D classes is depicted as insets. Scale bar, 50 nm. Negative staining for this sample was performed once, before advancing to cryo-EM analysis. d, Flowchart for cryo-EM analysis as described in Methods. e, 2D classification of particles reveal a 2D-crystalline arrangement of PhPINK1(D357A) oligomers in some areas of the grid. f, Final cryo-EM density maps (coloured by local resolution) for the PhPINK1(D357A) dodecamer (left) at 2.48 Å and the extracted dimer (right) at 2.35 Å. g, Examples of map quality for the final 2.35 Å density covering the PhPINK1(D357A) dimer.
Extended Data Fig. 4 Further analysis of the PhPINK1 oligomeric state.
a, Model built into the PhPINK1(D357A) dodecamer cryo-EM density as in Fig. 2b. b, Side view of the oligomer showing how the N-helix–CTR area facilitates contacts to connect four dimers. c, We were concerned that PhPINK1 oligomerization may only happen with a specific mutant protein and tested whether the oligomer would also form with unphosphorylated WT PhPINK1. WT PhPINK1 is phosphorylated at multiple sites when purified from E. coli (see10), and was hence dephosphorylated by λ-PP and repurified by SEC as indicated (see Methods). Like the inactive D357A mutant, a prominent oligomer, as well as a dimer/monomer equilibrium is apparent on SEC. Alternatively, the WT protein was first dephosphorylated by λ-PP, then rephosphorylated by adding Mg2+/ATP for 1 min, and the kinase and λ-PP were inactivated by EDTA (see Methods). Phosphorylation destabilized the oligomer to a predominant monomeric species, explaining why WT PhPINK1 does not form the oligomer, and suggesting that autophosphorylation resolves the oligomer and dimer (see below). These experiments were part of the purification process and were performed three times with identical results. d, SEC–MALS analysis of TcPINK1 variants, to show that corresponding TcPINK1 constructs (amino acids 117–570) do not show oligomeric behaviour. TcPINK1 purifications revealing monomeric behaviour were performed at least twice, and a SEC–MALS experiment was performed once. e, Residues mediating hydrogen bonds (dotted lines) in the oligomer interfaces. Lack of conservation of oligomer contacts between PhPINK1, TcPINK1 and HsPINK1 likely explain why TcPINK1 does not form an oligomer, and why we do not expect an identical oligomer in HsPINK1.
Extended Data Fig. 5 Structural and biochemical analysis of the PhPINK1 dimer.
a, Comparison between PhPINK1 autophosphorylation and ubiquitin phosphorylation resolved previously (PDB: 6EQI)10, with relevant details in the active site (top row) and in the activation segment (bottom row). The right panel shows substrate disposition in relation to a modelled ATP molecule as in Fig. 3b. The dimeric PhPINK1 autophosphorylation complex appears to place Ser202 in an ideal phospho-accepting position. b, Comparison of activation segment structures in all published PINK1 structures, revealing high structural similarity. Ser375 (PhPINK1)/Ser377 (TcPINK1) is shown in ball-and-stick representation; this residue was mutated to Asp in one prior structure (see Extended Data Fig. 1a). c, PhPINK1 Ser375 corresponds to Ser402 in HsPINK1, which is a reported phosphorylation site31,33, but is located within the activation segment and out of reach of the substrate-binding site within dimeric PhPINK1. Our structure does not reveal how autophosphorylation at this residue could be facilitated in cis or trans (left), nor how phosphorylation through e.g. an upstream kinase would contribute to PINK1 activity or function, since phosphorylation would likely disrupt the dimer (right). ATP was modelled as in Fig. 3b. d, Manual docking of dimeric PhPINK1 onto a cryo-EM structure of dimeric human TOM complex (PDB: 7CK6)61. The PhPINK1 dimer was oriented with its two N-helices (spanning ~80 Å) aligning with the two TOM7 subunits of the TOM complex dimer. TOM7 has been reported as essential for PINK1 stabilization on the TOM complex27,28. Note that some TOM components have considerable cytosolic domains that would need to be accommodated in addition to a PINK1 dimer. e, Unidentified density in the 2.35 Å cryo-EM map connects Cys169 in the dimer. f, Time course of PhPINK1 and TcPINK1 disulphide formation upon treatment with 2 mM H2O2, resolved on a non-reducing SDS–PAGE gel. The dimer/oligomer stabilizing PhPINK1 D357A mutation (Fig. 3) enables fast disulphide formation that is averted by an additional C169A mutation. TcPINK1 WT or D359A (equivalent to PhPINK1(D357A)) mutant do not show fast cross-linking behaviour observed in PhPINK1. Engineering of an additional Thr172 to Cys mutation (TcPINK1(T172C/D359A)) results in rapid emergence of cross-linked TcPINK1 dimers upon oxidation. Experiments were performed in biological triplicate with identical results. See Supplementary Fig. 1 for uncropped gels. g, EOPD mutations in the activation segment and αEF–αF loop. Mutations according to10. h, HsPINK1 EOPD mutants listed in g were expressed in HeLa PINK1−/− cells, stabilized with OA and treated with H2O2 to assess PINK1 dimerization, autophosphorylation and ubiquitin phosphorylation activity (see Methods). The control D384A mutant (mutation of the DFG motif, equivalent to D357A in PhPINK1) was included as an inactive mutant. As anticipated, it is able to dimerize (oxidative cross-link formed) but unable to autophosphorylate or generate phosphorylated ubiquitin. Oxidative dimerization enables separation of pure catalytic mutants and dimerization deficient mutants. Experiments were performed in biological triplicate with identical results. See Supplementary Fig. 1 for uncropped blots.
Extended Data Fig. 6 Analysis of autophosphorylation sites in PhPINK1 and HsPINK1.
a, Phos-tag analysis of inactive, monomeric PhPINK1(D334A), which does not autophosphorylate when incubated with ATP. Phosphorylation by WT GST-tagged PhPINK1 for 2 h leads to a shift of the entire protein consistent with a single phosphorylation site, and a small amount (1–2%) shifts to a higher species indicating additional autophosphorylation events after prolonged incubation. A representative gel of three independent experiments is shown. See Supplementary Fig. 1 for uncropped gel. b, As in a but shown in a time course experiment. Mutating Ser202 to Ala abrogates the observed gel shift, indicating that Ser202 is the sole site of phosphorylation. Mutation of Ser375 (equivalent to HsPINK1 Ser402) to Ala does not impede phosphorylation. A representative gel of three independent experiments is shown. See Supplementary Fig. 1 for uncropped gel. c, Mass spectrometry confirms the detection of a phosphate at Ser202 of PhPINK1(D334A) after phosphorylation with WT GST–PhPINK1. See Methods. d, Time course experiment as in b, with monomeric PhPINK1(D334A) or oligomeric PhPINK1(D357A). While the monomer can be phosphorylated by WT GST–PhPINK1, the oligomer is not efficiently phosphorylated since Ser202 is buried in the stable dimer/oligomer structure. This experiment was performed twice with identical results. See Supplementary Fig. 1 for uncropped gel. e, Time course experiment using dephosphorylated WT PhPINK1 (amino acids 119–575, which we found to be oligomerization impaired) and PhPINK1 (amino acids 119–575) S202A, and with ubiquitin in the reaction. See Methods for details. Phosphorylation of PhPINK1 at Ser202 is essential for ubiquitin phosphorylation, as both phosphorylation events are completely abrogated if Ser202 cannot be phosphorylated. Representative gels of three independent experiments are shown. See Supplementary Fig. 1 for uncropped gels. f–i, Analysis of autophosphorylation sites in HsPINK1. f, In HsPINK1, the Ser202 equivalent residue is Ser228, which is a known, important autophosphorylation site. In PhPINK1, the Ser202 site is followed by Asn203 and Ser204, and Ser204 can be phosphorylated when PhPINK1 is expressed in E. coli10. Also, the Ser204 equivalent residue in TcPINK1 (Ser207) was phosphorylated in a previous crystal structure (Extended Data Fig. 1a). In HsPINK1, Ser228 is followed by Ser229 and Ser230, and we investigated whether phosphorylation of these residues occurs and whether it contributes to HsPINK1 activity. (g) Structural detail of the Ser202-containing loop in the active site of the PhPINK1 autophosphorylation dimer. Equivalent HsPINK1 residues are in brackets. ATP was modelled as in Fig. 3b. Only Ser202 is in the phospho-acceptor site, but the other residues may occupy the site and be phosphorylated subsequently, if the end of the αC helix slightly unravels; since we expect conformational changes in this region (see below), this seemed feasible. h, i, HsPINK1 variants with combinations in the Ser228-containing loop as indicated (empty, vector control; KD, kinase dead, a triple mutant K219A, D362A (HRD motif), D384A (DFG motif); SSS (WT) refers to the WT sequence with Ser228, Ser229, Ser230; ASS refers to Ala228, Ser229, Ser230; etc.) were expressed in HeLa PINK1−/− cells, treated with OA for 2 h, and subjected to western blotting (see Methods). h, A Phos-tag gel probed with anti-PINK1 antibody reveals that KD and AAA mutants remain unphosphorylated, while the WT HsPINK1 (SSS) shows multiple phosphorylation states, indicating more than one phosphorylation event in this loop. All three Ser residues appear to be phosphorylatable. These results were correlated with appearance of Ser65-phosphorylated ubiquitin in the same cell lysates, revealed by a ubiquitin Ser65-phosphospecific antibody. WT HsPINK1 leads to a strong signal for phosphorylated ubiquitin, while Ala mutation in Ser228 does not lead to a phosphorylated ubiquitin signal. In contrast, if only Ser228 is present (e.g. SAA mutant) the phosphorylated ubiquitin signal is indistinguishable from that of WT HsPINK1. i, As in h, but with further substitution of Ser228. Phosphomimetic residues Asp228 or Glu228, followed by Ala229 and Ala230, lead to a small increase in phosphorylated ubiquitin, in the absence of autophosphorylation. The results in h and i indicate that also in HsPINK1, Ser228 phosphorylation is essential to turn PINK1 into a ubiquitin kinase, and a phosphomimetic is a weak substitute. Other residues in this area may also become phosphorylated but do not trigger ubiquitin phosphorylation. Experiments shown in h and i were performed in biological triplicate with identical results. See Supplementary Fig. 1 for uncropped blots.
Extended Data Fig. 7 Structure of a cross-linked, phosphorylated PhPINK1 reveals asymmetry and conformational changes in the N-lobe.
a, Final phosphorylated and cross-linked PhPINK1 oligomer on a non-reducing SDS–PAGE gel. See Supplementary Fig. 1 for uncropped gel. b, Flowchart for cryo-EM analysis, as in Extended Data Fig. 3. c, Local resolution maps and resolution calculations. d, Map quality for selected regions. e, EM density for the phosphorylated Ser202 of molecule A, in the substrate-binding site of molecule B.
Extended Data Fig. 8 Assessment of PhPINK1 dimer stability by measuring oligomer formation.
a, Our studies had revealed that disruption of the kinase–substrate interaction had a detrimental effect for dimer formation. We wondered whether the underlying dimerization mechanism would facilitate the reported effect that each copy of PINK1 is phosphorylated during mitophagy18,30,33. Based on our biochemical data (Fig. 2a and Extended Data Fig. 4c) we conceptualized PINK1 dimer interactions as a function of the number of intact Ser202–Asp334 contacts. Ser202 phosphorylation would resolve the Ser202–Asp334 contacts individually, leading to one contact point after the first phosphorylation event, and zero contact points when both PINK1 molecules are phosphorylated. It was unclear whether the dimer is stable after the first phosphorylation event, when only one Ser202–Asp334 contact point exists within the dimer. b, We explored whether we would be able to generate PhPINK1 oligomers with an unphosphorylated PhPINK1(D334A) mutant and Ser202-phosphorylated PhPINK1 (phospho-PhPINK1), as indicated, with the knowledge that homo-oligomerization is disfavoured (zero Ser202–Asp334 contact points, highlighted in red). Formation of a hetero-oligomer between unphosphorylated PhPINK1(D334A) and phospho-PhPINK1 would indicate that the dimer is stable with just one Ser202–Asp334 contact (highlighted in green). c, SEC analysis of the mutants confirms that unphosphorylated PhPINK1(D334A) or phospho-PhPINK1 do not form oligomers, and hence, zero Ser202–Asp334 contact points are not sufficient. In contrast, enabling one contact point, as per the green scenario in b where variants are mixed, enables oligomerization of PhPINK1. Each individual protein showed identical results in every purification run (n = 3), and the mixing experiments were performed in biological triplicate. d, Phos-tag analysis confirms that the oligomer species is a 1:1 mixture of unphosphorylated PhPINK1(D334A) and phospho-PhPINK1. Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped gel. Together, these experiments show that the dynamic equilibrium favours dimer formation until each copy of PINK1 has been phosphorylated, ensuring full phosphorylation.
Extended Data Fig. 9 Regulation of PINK1 activity by oxidation.
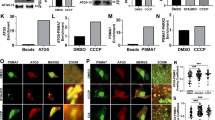
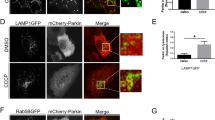
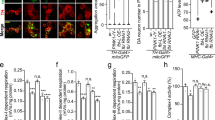
a, Extended sequence alignment indicating that Cys169 and Cys360 are well conserved in PINK1, and invariant in mammalian PINK1. Cys169 is a Thr in TcPINK1, and a Ser in many fish species. b, Comparison of PhPINK1 and TcPINK1, for their ability to be regulated by oxidation, shown in Phos-tag ubiquitin phosphorylation assays (see Fig. 5b). While PhPINK1 activity is abrogated with H2O2, TcPINK1 with Thr172 in the P-loop, remains active. The observed reduction in TcPINK1 activity could be a result of oxidation of the conserved Cys362 (Cys360 in PhPINK1) in the active site. Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped gel. c, Inhibition of PhPINK1 ubiquitin phosphorylation activity can be reversed with DTT, suggesting reversible regulatory oxidation. See Methods. Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped gel. d, Time course assessment of HsPINK1 Cys–Ala mutants transiently expressed in HeLa PINK1−/− cells. OA treatment leads to accumulation of HsPINK1 and slightly altered autophosphorylation for both C166A and C387A. The HsPINK1(C166A) mutant was almost completely deficient in ubiquitin phosphorylation, while HsPINK1(C387A) showed highly reduced but still detectable phosphorylated ubiquitin levels. Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped blots. e, Time course of HsPINK1 mutants as in d, but using stable HsPINK1 expression in the presence of YFP–Parkin. Presence of YFP–Parkin seemingly increases levels of phosphorylated ubiquitin for all HsPINK1 variants as compared to d, yet overall, phosphorylated ubiquitin and phosphorylated Parkin levels remain strongly diminished with HsPINK1(C166A), and to a lesser degree with HsPINK1(C387A). Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped blots. f, Translocation of YFP–Parkin (cyan) to mitochondria (magenta, TOM20–Halo) in HeLa PINK1−/− stably expressing HsPINK1 Cys–Ala variants upon OA treatment, imaged using lattice light sheet microscopy. Maximum intensity projections are shown for four different timepoints. YFP–Parkin translocation is delayed in cells expressing either HsPINK1(C166A) or HsPINK1(C387A) relative to WT hHsPINK1. A kinase dead (KD) HsPINK1 variant was included as a control. Images are representative of three independent experiments. Scale bar is 10 μm. See Supplementary Video 1. g, Quantification of YFP–Parkin translocation in f. The cumulative fraction of cells exhibiting YFP–Parkin translocation is shown over time. Approximately 70 cells were counted per cell line. Each curve was fitted to determine the time for 50% of the cells to feature translocation. Significant differences between curves were determined using a two-sample Kolmogorov–Smirnov test. A MATLAB script and Source Data are available as Supplementary Material. Exact p values are: WT–C166A: p < 2.83 × 10−20; WT–C387A: p < 2.36 × 10−18. h, HsPINK1 mutants expressed in HeLa PINK1−/− cells and treated with OA for 2 h. Activity of HsPINK1(C166A) and HsPINK1(C166S) can be partially restored by an additional S167N mutation, mimicking the sequence observed in many fish species. A TcPINK1-like sequence introduced into HsPINK1, C166T/S167N, is less active than the HsPINK1(C166S/S167N) mutant. The additional Asn in the P-loop of the HsPINK1(C166S/S167N) mutant recovers activity of HsPINK1(C166S), suggesting that both residues are also important in ubiquitin/Ubl substrate interactions, and not merely involved in dimerization. Experiments were performed in biological triplicate. See Supplementary Fig. 1 for uncropped blots. i, A redox active switch in fructosamine-3-kinases (PDB: 6OID) is conceptually similar, utilizing a Cys at an identical position (Cys32) for Cys-mediated cross-linking and regulation of kinase activity by oxidation36, however the overall orientation of kinase domains is dissimilar.
Extended Data Fig. 10 Analysis of the HsPINK1 model as predicted by AlphaFold237,38.
(a, b) Predicting the structure of HsPINK1 via AlphaFold2 (a, left), resulted in a model remarkably similar to PhPINK1 from the PhPINK1–Ub TVLN complex10 (b, right), and features a kinked αC helix, ordered insertion-3, and an extended N-helix that binds and extends from the CTR directly into the membrane. Consistently, predicting a complex between HsPINK1 with the ubiquitin-like (Ubl) domain of human Parkin (a, right), places the Ubl domain at HsPINK1 analogously to the PhPINK1–Ub TVLN complex (b, right). Both predictions are dissimilar to unphosphorylated PhPINK1 (b, left) and TcPINK1 (Extended Data Fig. 1). Insets show the detail of the N-lobe with a kinked αC helix and an ordered insertion-3. The prediction is somewhat surprising, as it suggests HsPINK1 to be an active ubiquitin kinase even without Ser228 phosphorylation, which contradicts biochemical analysis. Since AlphaFold2 does not yet predict the impact of post-translational modifications, we interpret the prediction such that it is possible and even likely, that HsPINK1 can adopt a ubiquitin-phosphorylation-competent conformation consistent with our previous PhPINK1–Ub TVLN complex structure10. c, d, We next used AlphaFold2 to predict a dimer of HsPINK1. c, Strikingly, AlphaFold2 predicts a symmetric dimer with a dimer interface identical to the one shown for PhPINK1 (compare with d and Fig. 3). In fact, we have already validated this arrangement of HsPINK1 molecules via our Cys166 cross-linking experiments in Fig. 3g. However, in the predicted dimer of HsPINK1, the αC helix is kinked and Ser228 does not contact the second molecule. This is different from our conclusions in Extended Data Fig. 8, but again may be a result of not incorporating the effect of Ser228 phosphorylation. We therefore anticipate that unphosphorylated HsPINK1 can also adopt a conformation with an extended αC helix that places Ser228 into the active site of the dimeric molecule to facilitate autophosphorylation prior to forming the depicted conformation. AlphaFold2 predictions hence support the notion that the activation model proposed in Fig. 5g applies to HsPINK1.
Supplementary information
Supplementary Figs. 1 and 2
Supplementary Fig. 1: uncropped gels and blots. Supplementary Fig. 2: repeats of Fig. 5e, used for quantification in Fig. 5f.
Supplementary Tables 1 and 2
Crystallographic and cryo-EM data.
Supplementary Data
Source code: MATLAB script to reproduce Kolmogorov–Smirnov test in Extended Data Fig. 9g.
Supplementary Video 1
YFP–Parkin translocation in PINK1−/− HeLa cells expressing HsPINK1 variants. Live cell imaging of YFP–Parkin (cyan) translocation to the mitochondria (magenta, TOM20–Halo) after OA treatment in PINK1−/− HeLa cells stably expressing the indicated HsPINK1 variants. KD, kinase dead. Compare with Extended Data Fig. 9. Images were taken every minute for 60 min using a Lattice Light Sheet 7 (Zeiss) microscope. Maximum intensity projections are shown for four different timepoints. Images are representative of three independent experiments. Scale bar, 10 μm. Frame rate, approximately 3 fps.
Rights and permissions
About this article
Cite this article
Gan, Z.Y., Callegari, S., Cobbold, S.A. et al. Activation mechanism of PINK1. Nature 602, 328–335 (2022). https://doi.org/10.1038/s41586-021-04340-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04340-2
This article is cited by
-
Identification and structural characterization of small molecule inhibitors of PINK1
Scientific Reports (2024)
-
The mitophagy pathway and its implications in human diseases
Signal Transduction and Targeted Therapy (2023)
-
The Role of Ubiquitin–Proteasome System and Mitophagy in the Pathogenesis of Parkinson's Disease
NeuroMolecular Medicine (2023)
-
Targeting mitophagy for neurological disorders treatment: advances in drugs and non-drug approaches
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
An increase in mitochondrial TOM activates apoptosis to drive retinal neurodegeneration
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.