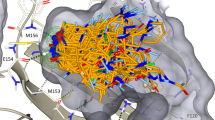
Abstract
Structure-based virtual ligand screening is emerging as a key paradigm for early drug discovery owing to the availability of high-resolution target structures1,2,3,4 and ultra-large libraries of virtual compounds5,6. However, to keep pace with the rapid growth of virtual libraries, such as readily available for synthesis (REAL) combinatorial libraries7, new approaches to compound screening are needed8,9. Here we introduce a modular synthon-based approach—V-SYNTHES—to perform hierarchical structure-based screening of a REAL Space library of more than 11 billion compounds. V-SYNTHES first identifies the best scaffold–synthon combinations as seeds suitable for further growth, and then iteratively elaborates these seeds to select complete molecules with the best docking scores. This hierarchical combinatorial approach enables the rapid detection of the best-scoring compounds in the gigascale chemical space while performing docking of only a small fraction (<0.1%) of the library compounds. Chemical synthesis and experimental testing of novel cannabinoid antagonists predicted by V-SYNTHES demonstrated a 33% hit rate, including 14 submicromolar ligands, substantially improving over a standard virtual screening of the Enamine REAL diversity subset, which required approximately 100 times more computational resources. Synthesis of selected analogues of the best hits further improved potencies and affinities (best inhibitory constant (Ki) = 0.9 nM) and CB2/CB1 selectivity (50–200-fold). V-SYNTHES was also tested on a kinase target, ROCK1, further supporting its use for lead discovery. The approach is easily scalable for the rapid growth of combinatorial libraries and potentially adaptable to any docking algorithm.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Chemical structures, synthetic methods, detailed results of biochemical characterization are presented in this paper and its Supplementary Information.
Code availability
V-SYNTHES scripts and example files have been deposited at GitHub (https://github.com/katritchlab/V-SYNTHES).
References
Shoichet, B. K. & Kobilka, B. K. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 33, 268–272 (2012).
Katritch, V., Cherezov, V. & Stevens, R. C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556 (2013).
Renaud, J.-P. et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 (2018).
Congreve, M., de Graaf, C., Swain, N. A. & Tate, C. G. Impact of GPCR structures on drug discovery. Cell 181, 81–91 (2020).
Stein, R. M. et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Grygorenko, O. O. et al. Generating multibillion chemical space of readily accessible screening compounds. iScience 23, 101681 (2020).
Gorgulla, C. et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020).
Graff, D. E., Shakhnovich, E. I. & Coley, C. W. Accelerating high-throughput virtual screening through molecular pool-based active learning. Chem. Sci. 12, 7866–7881 (2021).
Engels, M. F. & Venkatarangan, P. Smart screening: approaches to efficient HTS. Curr. Opin. Drug Discov. Dev. 4, 275–283 (2001).
Villoutreix, B. O., Eudes, R. & Miteva, M. A. Structure-based virtual ligand screening: recent success stories. Comb. Chem. High Throughput Screen. 12, 1000–1016 (2009).
Abagyan, R. & Totrov, M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 5, 375–382 (2001).
Irwin, J. J. & Shoichet, B. K. Docking screens for novel ligands conferring new biology. J. Med. Chem. 59, 4103–4120 (2016).
Ertl, P. Cheminformatics analysis of organic substituents: identification of the most common substituents, calculation of substituent properties, and automatic identification of drug-like bioisosteric groups. J. Chem. Inf. Comput. Sci. 43, 374–380 (2003).
Bohacek, R. S., McMartin, C. & Guida, W. C. The art and practice of structure-based drug design: a molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996).
REAL Space (Enamine, 2020); https://enamine.net/library-synthesis/real-compounds/real-space-navigator
Guzmán, M. Cannabinoids: potential anticancer agents. Nat. Rev. Cancer 3, 745–755 (2003).
Contino, M., Capparelli, E., Colabufo, N. A. & Bush, A. I. Editorial: the CB2 cannabinoid system: a new strategy in neurodegenerative disorder and neuroinflammation. Front. Neurosci. 11, 196 (2017).
Lunn, C. A. et al. Biology and therapeutic potential of cannabinoid CB2 receptor inverse agonists. Br. J. Pharmacol. 153, 226–239 (2008).
Corey, E. J. General methods for the construction of complex molecules. Pure Appl. Chem. 14, 19–38 (1967).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Li, X. et al. Crystal structure of the human cannabinoid receptor CB2. Cell 176, 459–467 (2019).
Kroeze, W. K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012).
Xing, C. et al. Cryo-EM structure of the human cannabinoid receptor CB2-Gi signaling complex. Cell 180, 645–654 (2020).
Wei, L., Surma, M., Shi, S., Lambert-Cheatham, N. & Shi, J. Novel insights into the roles of rho kinase in cancer. Arch. Immunol. Ther. Exp. 64, 259–278 (2016).
Chin, V. T. et al. Rho-associated kinase signalling and the cancer microenvironment: novel biological implications and therapeutic opportunities. Expert Rev. Mol. Med. 17, e17 (2015).
Baker, M. Fragment-based lead discovery grows up. Nat. Rev. Drug Discov. 12, 5–7 (2013).
Schulz, M. N. & Hubbard, R. E. Recent progress in fragment-based lead discovery. Curr. Opin. Pharmacol. 9, 615–621 (2009).
Davis, B. J. & Hubbard, R. E. in Structural Biology in Drug Discovery 79–98 (2020).
Zheng, Z. et al. Structure-based discovery of new antagonist and biased agonist chemotypes for the kappa opioid receptor. J. Med. Chem. 60, 3070–3081 (2017).
de Graaf, C. et al. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H1 receptor. J. Med. Chem. 54, 8195–8206 (2011).
Katritch, V. et al. Structure-based discovery of novel chemotypes for adenosine A2A receptor antagonists. J. Med. Chem. 53, 1799–1809 (2010).
Chen, Y. & Shoichet, B. K. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nat. Chem. Biol. 5, 358–364 (2009).
Abagyan, R. A., Orry, A., Raush, E., Budagyan, L. & Totrov, M. ICM User’s Guide and Reference Manual v.3.9 (MolSoft, 2021).
Bogolubsky, A. V. et al. A one-pot parallel reductive amination of aldehydes with heteroaromatic amines. ACS Comb. Sci. 16, 375–380 (2014).
Savych, O. et al. One-pot parallel synthesis of 5-(dialkylamino)tetrazoles. ACS Comb. Sci. 21, 635–642 (2019).
Katritch, V., Rueda, M. & Abagyan, R. Ligand-guided receptor optimization. Methods Mol. Biol. 857, 189–205 (2012).
Gatica, E. A. & Cavasotto, C. N. Ligand and decoy sets for docking to G protein-coupled receptors. J. Chem. Inf. Model. 52, 1–6 (2012).
Bottegoni, G., Kufareva, I., Totrov, M. & Abagyan, R. Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J. Med. Chem. 52, 397–406 (2009).
Real Compound Libraries (Enamine, 2020); https://enamine.net/library-synthesis/real-compounds/real-compound-libraries
Nikas, S. P. et al. Probing the carboxyester side chain in controlled deactivation (−)-Δ8-tetrahydrocannabinols. J. Med. Chem. 58, 665–681 (2015).
Nikas, S. P. et al. Novel 1′,1′-chain substituted hexahydrocannabinols: 9β-hydroxy-3-(1-hexyl-cyclobut-1-yl)-hexahydrocannabinol (AM2389) a highly potent cannabinoid receptor 1 (CB1) agonist. J. Med. Chem. 53, 6996–7010 (2010).
Jacobs, M. et al. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J. Biol. Chem. 281, 260–268 (2006).
Anastassiadis, T., Deacon, S. W., Devarajan, K., Ma, H. & Peterson, J. R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1039–1045 (2011).
Acknowledgements
We thank the staff at the USC Center for Advanced Research Computing, and the Google Cloud Platform for Higher Education and Research for providing computational resources. The study was funded by National Institute on Drug Abuse grants R01DA041435 and R01DA045020 (to V.K. and A.M.), National Institute of Mental Health Grant R01MH112205 and Psychoactive Drug Screening Program (to B.L.R.) and the Michael Hooker Distinguished Professorship (to B.L.R.). B.H. was supported by NIGMS T32-GM118289.
Author information
Authors and Affiliations
Contributions
A.A.S. and A.V.S. developed V-SYNTHES algorithms, performed calculations and wrote the first draft of the manuscript. B.H. and N.A.P. performed calculations and compound selection for ROCK1. Y.L., M.K.J., J.P. and X.-P.H. performed functional and selectivity assays. C.I.-T., N.K.T., F.T., N.Z. and S.P.N. performed binding assays. N.P. performed full VLS on Google Cloud. O.S., D.S.R. and Y.S.M. developed the REAL Space library and performed compound synthesis. B.L.R. supervised the functional and selectivity assays. A.M. supervised binding assays for CB1 and CB2. V.K. conceived the study and supervised all of its computational aspects. All of the authors contributed to writing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.A.S. and V.K. filed a provisional patent on V-SYNTHES method (application no. 63159888, University of Southern California).
Additional information
Peer review information Nature thanks Charlotte Dean and Amy Newman for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Evaluation of SYNTHES performance on CB2 receptor with only docking score (without considering docking pose of MEL candidates in the binding pocket).
(a) The number of hits at each score threshold from V-SYNTHES and standard VLS (b) Enrichment in V-SYNTHES vs. Standard VLS at different score thresholds, with the red x-mark showing threshold that yields 100 V-SYNTHES hits in the two-component library.
Extended Data Fig. 2 Binding pocket of CB2 with selected dead-end atoms.
a) 3D illustration of a MEL compound binding pose (carbon atoms colored cyan) with a “non-productive” pose. (b-d) 2D schematics showing other possible non-productive cases, including dead-end subpockets. Dead-end water-colored red, pseudoatoms colored magenta.
Extended Data Fig. 3 Details of practical application V-SYNTHES algorithms to CB receptors screening.
a, b, Two-component (a) and three-component (b) reaction cases.
Extended Data Fig. 4 Concentration-response curves for V-SYNTHES hits in functional assays at CB1 and CB2 receptors (except those shown in main text Figure 3).
β-arrestin recruitment Tango assays were performed to assess antagonist activity of the compounds in (a,b) CB1 and (c,d) CB2 receptors. The compounds rimonabant or SR144528 served as positive controls. The assays were carried out in the presence of 100 nM (EC80) of the dual CB1/CB2 CP55,940 agonist. The data points are presented as mean ± SEM with n = 3 independent experiments, each one carried out in triplicate.
Extended Data Fig. 5 Competition binding curves for the best CB2 hit compounds from V-SYNTHES.
Radioligand binding assays were used to assess the binding affinities in rCB1 (a) and hCB2 (b). [3H]CP-55,940 was used as the radioligand. The data were presented as mean ± SEM with n = 3 independent experiments, each one carried out in triplicate.
Extended Data Fig. 6 Assessment of off-target selectivity for the best V-SYNTHES CB2 hits.
(a-c) Screening of compounds 673, 610 and 523 at 10 µM concentrations in GPCRome-Tango assays for >300 receptors. Dopamine D2 (DRD2) and 100 nM Quinpirole served as an assay control. The data are presented as mean ± SEM (n = 4) and the values of fold of basal > 3 are marked as significant hits. (d-o) Follow-up dose-response curves for targets with >3 fold increased activity. Known agonists or antagonist that showed activity served as positive controls. The data points are presented as mean ± SEM with n = 3 independent experiments, each assay carried out in triplicate.
Extended Data Fig. 7 Identification and characterization of CB1 and CB2 hits from standard VLS of 115M Enamine REAL compounds.
(a) Chemical structures of the hits from the standard VLS. (b-c). Concentration-response curves of the best hits in β-arrestin recruitment Tango assays for antagonist activity at CB1 (b) and CB2 (c) receptors. The compounds rimonabant or SR144528 served as positive controls. The assays were carried out in the presence of 100 nM (EC80) of the dual CB1/CB2 CP55,940 agonist. The data points are presented as mean ± SEM with n = 3 independent experiments, each one carried out in triplicate. (d) Functional potencies and binding affinities of the hit compounds from standard VLS. The 95% Confidence Intervals (CI) were calculated from n = 3 independent assays, with 16 dose-response points for functional Ki values and 8 dose-response points for affinity Ki values, except for values marked with *, roughly estimated from three-point assays.
Extended Data Fig. 8 Competition binding curves for the best CB2 hit compounds from standard VLS.
Radioligand binding assays were used to assess the binding affinities in hCB2. [3H]CP-55,940 was used as the radioligand. The data were presented as mean ± SEM with n = 3 independent experiments, each one carried out in triplicate.
Extended Data Fig. 9 Chemical structures for series of the SAR-by-catalog analogues of antagonists, discovered by V-SYNTHES.
Shown are 60 analogues of 523 (a), 610 (b), and 673 (c) with inhibitory activity >40% in the single point functional assays. All 104 analogues tested are shown in Supplementary Information Table S3.
Extended Data Fig. 10 Functional potency and binding affinity assessment of the SAR-by-catalog analogues of the antagonist 523, discovered by V-SYNTHES.
Table compounds with CB2 potency better than 500 nM are shown, antagonists with affinities better than 10 nM highlighted in bold, >50-fold selective by italic. Functional Ki values and 95% Confidence Intervals were calculated from n = 4 independent assays with 16 dose-response points. Affinity Ki values and 95% Confidence Intervals were calculated from n = 3 independent assays with 8 dose-response points.
Extended Data Fig. 11 Concentration-response curves for series of the SAR-by-catalog analogues of 523, 610 and 673 antagonists, discovered by V-SYNTHES.
The β-arrestin recruitment Tango assays were performed to assess the antagonist activity of the best hits at CB1 (a-i), and CB2 (j-o) receptors. Note that the six best analogues of 523 shown in Fig. 4 are excluded here. The compounds rimonabant and SR144528 served as positive controls. The assays were carried out in the presence of 100 nM (EC80) of the CP55,940 agonist. The data were presented as mean ± SEM with n = 3 independent experiments, each run carried out in triplicate.
Extended Data Fig. 12 Assessment of off-target selectivity for the best SAR-by-catalog compounds 733 and 747.
(a-b) Screening of compounds 733 and 747 in GPCRome-Tango assay for >300 receptors at 10 µM concentrations. Dopamine D2 (DRD2) and 100 nM Quinpirole served as an assay control. The data are presented as mean ± SEM (n = 4) and the values of fold of basal > 3 marked as significant hits. (c-d) Follow-up dose-response curves for targets with >3 fold increased activity. Known agonists that showed activity served as positive controls. The data were presented as mean ± SEM with n = 3 independent experiments, each run carried out in triplicate.
Extended Data Fig. 13 Application of V-SYNTHES to the discovery of ROCK1 inhibitors.
(a,b) Computational assessment of V-SYNTHES performance vs standard VLS. (a) The number of candidate hits at each score threshold from V-SYNTHES and standard VLS. (b) Enrichment in V-SYNTHES vs. standard VLS at different score thresholds, with the red x-mark showing threshold that yields 100 hits in the two-component library. (c) Chemical structures of all selected by V-SYNTHES and synthesized compounds for ROCK1 kinase.
Extended Data Fig. 14 Experimental characterization of candidate ROCK1 inhibitors predicted by V-SYNTHES.
Full dose-response curves for the ROCK1 hits in (a) functional potency and (b) binding affinity at human ROCK1. The data points are presented as mean ± SEM from n = 3 independent experiments, each run carried out in triplicate. (c) Values of binding affinities and functional potencies for all candidate compounds predicted by V-SYNTHES. Bold font highlight hits with IC50<10 µM. Estimated values for curves that did not allow accurate fitting are marked with *.
Extended Data Fig. 15 Examples of typical Enamine REAL reactions.
(a) two-component reaction (b) three-component reaction.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Supplementary Tables 1–4.
Supplementary File 2
Detailed synthesis protocol for all compounds in the paper.
Supplementary File 3
NMR and LC–MS spectra for all compounds in the paper.
Supplementary File 4
HRMS spectra for all compounds in the paper.
Rights and permissions
About this article
Cite this article
Sadybekov, A.A., Sadybekov, A.V., Liu, Y. et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature 601, 452–459 (2022). https://doi.org/10.1038/s41586-021-04220-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04220-9
This article is cited by
-
Exploring the combinatorial explosion of amine–acid reaction space via graph editing
Communications Chemistry (2024)
-
A divergent intermediate strategy yields biologically diverse pseudo-natural products
Nature Chemistry (2024)
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
Generative AI for designing and validating easily synthesizable and structurally novel antibiotics
Nature Machine Intelligence (2024)
-
GraphsformerCPI: Graph Transformer for Compound–Protein Interaction Prediction
Interdisciplinary Sciences: Computational Life Sciences (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.