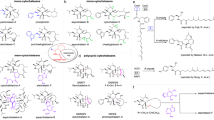
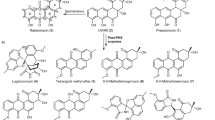
Abstratct
β-Nicotinamide adenine dinucleotide (β-NAD) is a pivotal metabolite for all living organisms and functions as a diffusible electron acceptor and carrier in the catabolic arms of metabolism1,2. Furthermore, β-NAD is involved in diverse epigenetic, immunological and stress-associated processes, where it is known to be sacrificially utilized as an ADP-ribosyl donor for protein and DNA modifications, or the generation of cell-signalling molecules3,4. Here we report the function of β-NAD in secondary metabolite biosynthetic pathways, in which the nicotinamide dinucleotide framework is heavily decorated and serves as a building block for the assembly of a novel class of natural products. The gatekeeping enzyme of the discovered pathway (SbzP) catalyses a pyridoxal phosphate-dependent [3+2]-annulation reaction between β-NAD and S-adenosylmethionine, generating a 6-azatetrahydroindane scaffold. Members of this novel family of β-NAD-tailoring enzymes are widely distributed in the bacterial kingdom and are encoded in diverse biosynthetic gene clusters. The findings of this work set the stage for the discovery and exploitation of β-NAD-derived natural products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data that support the findings of this study are available within the paper and its Supplementary Information, or are available from the corresponding authors upon request.
References
Walsh, C. T. & Tang, Y. The Chemical Biology of Human Vitamins (RSC, 2019).
Cantó, C., Menzies, K. J. & Auwerx, J. NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015).
Depaix, A. & Kowalska, J. NAD analogs in aid of chemical biology and medicinal chemistry. Molecules 24, 4187 (2019).
Schuller, M. et al. Molecular basis for DarT ADP-ribosylation of a DNA base. Nature 596, 597–602 (2021).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020).
Walsh, C. T. & Tang, Y. Natural Product Biosynthesis: Chemical Logic and Enzymatic Machinery (RSC, 2017).
Blin, K., Kim, H. U., Medema, M. H. & Weber, T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 20, 1103–1113 (2018).
Houge-Frydrych, C. S. V., Gilpin, M. L., Skett, P. W. & Tyler, J. W. SB-203207 and SB-203208, two novel isoleucyl tRNA synthetase inhibitors from a Streptomyces sp. II. Structure determination. J. Antibiot. 53, 364–372 (2000).
Stefanska, A. L., Cassels, R., Ready, S. J. & Warr, S. R. SB-203207 and SB-203208, two novel isoleucyl tRNA synthetase inhibitors from a Streptomyces sp. I. Fermentation, isolation and properties. J. Antibiot. 53, 357–363 (2000).
Takahashi, A., Kurasawa, S., Ikeda, D., Okami, Y. & Takeuchi, T. Altemicidin, a new acaricidal and antitumor substance. I. Taxonomy, fermentation, isolation and physico-chemical and biological properties. J. Antibiot. 42, 1556–1561 (1989).
Takahashi, A., Kurasawa, S., Ikeda, D., Okami, Y. & Takeuchi, T. Altemicidin, a new acaricidal and antitumor substance. II. Structure determination. J. Antibiot. 42, 1562–1566 (1989).
Yan, Y., Liu, N. & Tang, Y. Recent developments in self-resistance gene directed natural product discovery. Nat. Prod. Rep. 37, 879–892 (2020).
Hu, Z., Awakawa, T., Ma, Z. & Abe, I. Aminoacyl sulfonamide assembly in SB-203208 biosynthesis. Nat. Commun. 10, 184 (2019).
Bull, J. A., Mousseau, J. J., Pelletier, G. & Charette, A. B. Synthesis of pyridine and dihydropyridine derivatives by regio- and stereoselective addition to n-activated pyridines. Chem. Rev. 112, 2642–2713 (2012).
Eliot, A. C. & Kirsch, J. F. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415 (2004).
Du, Y. L. & Ryan, K. S. Pyridoxal phosphate-dependent reactions in the biosynthesis of natural products. Nat. Prod. Rep. 36, 430–457 (2019).
Cook, P. D. & Holden, H. M. A structural study of GDP-4-keto-6-deoxy-d-mannose-3-dehydratase: caught in the act of geminal diamine formation. Biochemistry 46, 14215–14224 (2007).
Hirayama, A., Miyanaga, A., Kudo, F. & Eguchi, T. Mechanism-based trapping of the quinonoid intermediate by using the K276R mutant of PLP-dependent 3-aminobenzoate synthase PctV in the biosynthesis of pactamycin. ChemBioChem 16, 2484–2490 (2015).
Harmange Magnani, C. S. & Maimone, T. J. Dearomative synthetic entry into the altemicidin alkaloids. J. Am. Chem. Soc. 143, 7935–7939 (2021).
Chen, M., Liu, C. T. & Tang, Y. Discovery and biocatalytic application of a PLP-dependent amino acid g-substitution enzyme that catalyzes C-C bond formation. J. Am. Chem. Soc. 142, 10506–10515 (2020).
Hai, Y., Chen, M., Huang, A. & Tang, Y. Biosynthesis of mycotoxin fusaric acid and application of a PLP-dependent enzyme for chemoenzymatic synthesis of substituted l-pipecolic acids. J. Am. Chem. Soc. 142, 19668–19677 (2020).
Spiering, M. J., Moon, C. D., Wilkinson, H. H. & Schardl, C. L. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169, 1403–1414 (2005).
Cui, Z. et al. Pyridoxal-5′-phosphate-dependent alkyl transfer in nucleoside antibiotic biosynthesis. Nat. Chem. Biol. 16, 904–911 (2020).
Brzovic, P. et al. Reaction mechanism of Escherichia coli cystathionine γ-synthase: direct evidence for a pyridoxamine derivative of vinylglyoxylate as a key intermediate in pyridoxal phosphate dependent γ-elimination and γ-replacement reactions. Biochemistry 29, 442–451 (1990).
Bailey, H. J. et al. Human aminolevulinate synthase structure reveals a eukaryotic-specific autoinhibitory loop regulating substrate binding and product release. Nat. Commun. 11, 1–12 (2020).
Raboni, S. et al. in Comprehensive Natural Products II: Chemistry and Biology Vol. 7 (eds Mander, L. & Liu, H.-W.) 273–315 (Elsevier, 2010).
Poulos, T. L. Heme enzyme structure and function. Chem. Rev. 114, 3919–3962 (2014).
Ruszczycky, M. W., Zhong, A. & Liu, H. W. Following the electrons: peculiarities in the catalytic cycles of radical SAM enzymes. Nat. Prod. Rep. 35, 615–621 (2018).
Mehta, A. P. et al. Radical S-adenosylmethionine (SAM) enzymes in cofactor biosynthesis: a treasure trove of complex organic radical rearrangement reactions. J. Biol. Chem. 290, 3980–3986 (2015).
Walsh, C. T. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 33, 127–135 (2016).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 1–8 (2008).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Roy, A., Yang, J. & Zhang, Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, 471–477 (2012).
Bashiri, G., Rehan, A. M., Greenwood, D. R., Dickson, J. M. J. & Baker, E. N. Metabolic engineering of cofactor F420 production in Mycobacterium smegmatis. PLoS ONE 5, 1–10 (2010).
Bashiri, G., Squire, C. J., Baker, E. N. & Moreland, N. J. Expression, purification and crystallization of native and selenomethionine labeled Mycobacterium tuberculosis FGD1 (Rv0407) using a Mycobacterium smegmatis expression system. Protein Expr. Purif. 54, 38–44 (2007).
Oyugi, M. A., Bashiri, G., Baker, E. N. & Johnson-winters, K. Investigating the reaction mechanism of F 420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium tuberculosis: kinetic analysis of the wild-type and mutant enzymes. Biochemistry 55, 5566–5577 (2016).
McCarty, R. M., Krebs, C. & Bandarian, V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry 52, 188–198 (2013).
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (JSPS KAKENHI grant numbers JP16H06443, JP17H04763, JP19H04641, JP20H00490, JP20KK0173 and JP21K18246), the New Energy and Industrial Technology Development Organization (NEDO, Grant Number JPNP20011) and AMED (Grant Number JP21ak0101164), Japan Science and Technology Agency (JST SICORP grant number JPMJSC1701), UTEC-UTokyo FSI Research Grant Program, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Takeda Science Foundation, Kato Memorial Bioscience Foundation, The Asahi Glass Foundation, and a Sir Charles Hercus Fellowship (Health Research Council of New Zealand) to G.B. (17/058). We thank GlaxoSmithKline for providing Streptomyces sp. NCIMB 40513; G. Cao for providing S. gilvosporeus F607; M. Kobayashi for providing pHSA81; Deutsche Forschungsgemeinschaft (DFG BA 6870/1-1) for a postdoctoral research fellowship to L.B.; T. Mori and R. Ushimaru for critical reading of the manuscript; F. Kudo, T. Eguchi and T. Ueda for instruction on the stopped flow apparatus; and H. Zhang, F. Hayashi and Y. Ishii for measurement of NMR spectra at RIKEN Yokohama.
Author information
Authors and Affiliations
Contributions
T.A. and I.A. conceived the idea for the study. L.B., T.A. and I.A. developed the hypothesis and designed the experiments. L.B. and T.A. performed the in vivo and in vitro experiments. L.B. and K.S. performed compound isolation and characterization. L.B. and T.A. performed bioinformatic analysis to identify biosynthetic gene clusters and protein functions. L.B., T.A. and K.S. performed protein purification. G.B. expressed SbzF and provided the F420 cofactor regeneration system. T.A. and Z.H. performed the feeding experiments. All authors analysed and discussed the results. L.B., T.A. and I.A. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Identification of the gatekeeping enzyme SbzP.
a, LC-MS analysis of single gene expression culture extracts (UV trace 340 nm). b, Summary of isotope feeding experiment results and retrobiosynthetic considerations. c, LC-MS analysis of the productive SbzP substrate combination of β-NAD (5) and SAM (6) with concomitant appearance of dinucleotide product 7 (UV trace 258 nm). “Control” indicates incubation of substrates without enzyme.
Extended Data Fig. 2 Gene deletion-guided in vitro reconstitution of the sbz pathway.
a, LC-MS analysis of gene deletion strain culture extracts (UV trace 280 nm). b, LC-MS analysis of SbzQ-mediated conversion of 7 to 8 (EIC traces of substrate and product). c, LC-MS analysis of SbzI reaction of 8 to 9 (UV trace 340 nm). d, LC-MS analysis of deadenosylribosylation catalyzed by SbzN+O+H (EIC traces of substrate and product). e, LC-MS analysis of F420-dependent reduction of 11 to 12 (EIC traces of substrate and products). f, LC-MS analysis of SbzE-mediated methylation towards 1 (UV trace 287 nm). g, Proposed steps in the deadenosylribosylation reaction. “Control” indicates incubation of substrates without enzyme(s).
Extended Data Fig. 3 SbzP as a functionally and phylogenetically distinct PLP-dependent enzyme family.
a, Time-resolved photospectroscopic changes of SbzP after addition of SAM (6) (see Supplementary Note 5 for further details). b, Phylogenetic analysis of SbzP and reported subfamily members of the AAT-I protein superfamily.
Supplementary information
Supplementary Information
This file contains Supplementary Text; Supplementary Figs. 1 – 81; Supplementary Table 1; Supplementary Notes 1–8 and Supplementary References
Rights and permissions
About this article
Cite this article
Barra, L., Awakawa, T., Shirai, K. et al. β-NAD as a building block in natural product biosynthesis. Nature 600, 754–758 (2021). https://doi.org/10.1038/s41586-021-04214-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04214-7
This article is cited by
-
Genome mining for unknown–unknown natural products
Nature Chemical Biology (2023)
-
Molecular basis for carrier protein-dependent amide bond formation in the biosynthesis of lincosamide antibiotics
Nature Catalysis (2023)
-
A pyridoxal 5′-phosphate-dependent Mannich cyclase
Nature Catalysis (2023)
-
Emerging mechanistic insights into the regulation of specialized metabolism in plants
Nature Plants (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.