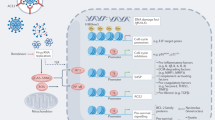
Abstract
Derailed cytokine and immune cell networks account for the organ damage and the clinical severity of COVID-19 (refs. 1,2,3,4). Here we show that SARS-CoV-2, like other viruses, evokes cellular senescence as a primary stress response in infected cells. Virus-induced senescence (VIS) is indistinguishable from other forms of cellular senescence and is accompanied by a senescence-associated secretory phenotype (SASP), which comprises pro-inflammatory cytokines, extracellular-matrix-active factors and pro-coagulatory mediators5,6,7. Patients with COVID-19 displayed markers of senescence in their airway mucosa in situ and increased serum levels of SASP factors. In vitro assays demonstrated macrophage activation with SASP-reminiscent secretion, complement lysis and SASP-amplifying secondary senescence of endothelial cells, which mirrored hallmark features of COVID-19 such as macrophage and neutrophil infiltration, endothelial damage and widespread thrombosis in affected lung tissue1,8,9. Moreover, supernatant from VIS cells, including SARS-CoV-2-induced senescence, induced neutrophil extracellular trap formation and activation of platelets and the clotting cascade. Senolytics such as navitoclax and a combination of dasatinib plus quercetin selectively eliminated VIS cells, mitigated COVID-19-reminiscent lung disease and reduced inflammation in SARS-CoV-2-infected hamsters and mice. Our findings mark VIS as a pathogenic trigger of COVID-19-related cytokine escalation and organ damage, and suggest that senolytic targeting of virus-infected cells is a treatment option against SARS-CoV-2 and perhaps other viral infections.
Similar content being viewed by others
Main
The causative agent of the COVID-19 pandemic, the human pathogenic coronavirus SARS-CoV-2, causes upper respiratory infections and subsequently lung disease that may become further complicated by septic multiorgan failure and is associated with increased mortality10,11. Escalating immune activation with massive cytokine release seems to drive severe COVID-19 disease1,2,3, possibly more than the virus infection itself. Mechanisms of viral entry—primarily through the interaction of the SARS-CoV-2 spike (S) protein with the host cell receptor angiotensin-converting enzyme 2 (ACE2)—and consecutive steps of RNA replication, virus assembly and budding represent therapeutic targets12,13. Anti-inflammatory drugs are clinically being used or currently being tested; however, they have led to mixed results14,15. Therefore, more comprehensive and specific measures to control the cytokine storm are needed.
Viral entry reflects a major cellular insult and induces marked biological changes in infected host cells that potentially lead to cellular senescence as a virus-triggered state switch16,17. This can have a disparate impact on virus propagation18,19. Senescence is also acutely stress-induced by activated oncogenes (termed oncogene-induced senescence (OIS)) or by DNA-damaging cancer therapy, and physiologically occurs in embryonic development, wound healing and ageing6. Little is known about systemic ramifications of cellular senescence as an antiviral host defence. We hypothesize here that cellular senescence and its SASP, which is largely composed of pro-inflammatory cytokines, matrix-degrading proteases and pro-coagulatory factors5,6,7, underlie the cytokine storm, macrophage activation, the formation of neutrophil extracellular traps (NETs), endothelialitis and widespread pulmonary thrombosis frequently observed in patients with COVID-19 (refs. 1,8,9).
Properties of VIS
We first tested signs of senescence in human diploid fibroblast (HDF) models exposed to high-titre retrovirus. At day 5 after infection, most of the cells presented with characteristics typical of senescence. The cells had flattened, enlarged morphology and were stained positive in the senescence-associated β-galactosidase (SA-β-gal) assay20. They also exhibited 4′,6-diamidino-2-phenylindole (DAPI)-dense chromatin reminiscent of senescence-associated heterochromatin foci (SAHF)21 and overexpressed p16INK4A, which is an inhibitor of cyclin-dependent kinase 4 (CDK4) and CDK6, in the nucleus (Fig. 1a, Extended Data Fig. 1a). Gene set enrichment analysis (GSEA) of HDF RNA sequencing (RNA-seq) datasets showed that senescence- and SASP-related Gene Ontology terms were skewed towards the VIS state and overlapped with the OIS state of the same cells in Ras-induced senescence (Fig. 1b). GSEA also retrieved host responses to viral infection in OIS, which further underlines the overarching role of a senescent stress response in this context (Extended Data Fig. 1b). Specific analyses of VIS-associated expression changes by quantitative PCR with reverse transcription (RT–qPCR) showed transcriptional changes typical of senescence in VIS cells21,22 (Fig. 1c, Extended Data Fig. 1c). Notably, lower virus titres did not induce senescent cell cycle arrest, which is the goal in retrovirus- or lentivirus-assisted gene transfer experiments (Extended Data Fig. 1d). Multiplex protein analysis showed that VIS cells secreted a plethora of SASP factors, many of which are bona fide NF-κB targets23,24 (Fig. 1d). Importantly, these responses depended on gene activities that are essential in senescence. For example, HDFs expressing the H3K9me3-active demethylase JMJD2C or engineered with a small hairpin RNA (shRNA) to stably knockdown p53 (also known as TP53) to genetically disable OIS22,25, and mouse embryo fibroblasts without intact p53 alleles failed to enter VIS and lacked senescence-typical changes, as assessed using RT–qPCR and multiplex secretome analyses (Fig. 1e, Extended Data Fig. 1e–g). After viral infection, only VIS-capable cells showed activation of the cGAS–STING pathway (Extended Data Fig. 1h), which is a cytoplasmic DNA sensor driving SASP-mediated interferon responses26. Notably, blocking retroviral replication using the reverse transcriptase inhibitor azidothymidine prevented VIS and blunted cGAS–STING pathway activation (Extended Data Fig. 1i, j). cGAS–STING-mediated phospho-activation of the interferon regulatory factor IRF3 (ref. 27), the NF-κB transcription factor p65 (ref. 28) and other nuclear senescence markers, such as p21CIP1 and heterochromatin-associated H3K9me3, were strongly increased in VIS cells. Conversely, non-G1-phase-hyperphosphorylated retinoblastoma (Rb) protein was markedly decreased in these cells (Fig. 1f). Hence, viral infection induces a host cell response reminiscent of and genetically dependent on key features of other well-established types of cellular senescence and their associated secretome.
a, SA-β-gal, SAHF formation by DAPI and p16INK4A staining of IMR90 cells 5 days after retroviral infection or mock control. b, GSEA probing senescence-associated genes in WI38 HDFs in VIS and OIS. A positive normalized enrichment score (NES) indicates enrichment compared to proliferating counterparts. Presented NES are significant (false discovery rate (FDR) q < 0.05; see Supplementary Information for individual values). c, Relative expression levels of core senescence and SASP transcripts in cells as in a by RT–qPCR compared to mock control. d, Multiplex bead-based protein analysis in the supernatant of HDF IMR90 (as in a), WI38 and Tig3 cells. MIP-1α is also known as CCL3; MIP-1β is also known as CCL4. e, SA-β-gal staining of IMR90 cells expressing JMJD2C, p53 shRNA (shp53) or control vector. f, Immunoblot analysis of nuclear senescence markers in samples as in d (α-tubulin was used as a loading control). g, Quantification of SA-β-gal positivity in human cell lines (RPE1, A549) infected with AAV, lentivirus, HCoV-NL63 or VSV. h, Quantification of SA-β-gal positivity in human cell lines and primary HNEpC cells infected with VSV-ΔG*-CoV-S, VSV-ΔG*-CoV-2-S or VSV-ΔG*/empty vector (VSV-ΔG*-emp). NIH3T3 cells were used as ACE2-negative, infection-resistant controls. i, SA-β-gal staining of stable hACE2-engineered primary nasal epithelial cells (HNEpC-hACE2) infected with SARS-CoV-2 and treated as indicated at 48 h after infection. GS, GS-441425; UT, untreated. j, Relative expression levels of the indicated transcripts by RT–qPCR of cells as in i, k, GSEA of RNA-seq-based GEP from AECs infected with SARS-CoV-2 compared with uninfected controls (n = 4). Preselected senescence-associated terms with FDR q ≤ 0.01 are shown (see Supplementary Information for individual values). Scale bars, 100 µm (a, e) or 5 µm (SAHF row in a). All quantifications in this figure refer to the mean ± s.d. of n = 3 independent experiments (except k) with individual values as dots in bar plots or representative photomicrographs shown.
Notably, VIS became detectable in response to a broad spectrum of viruses, including lentivirus, adeno-associated virus (AAV), vesicular stomatitis virus (VSV) and the low-pathogenic human alphacoronavirus NL63 (HCoV-NL63) in human lung carcinoma cells and in non-malignant epithelial cells (Fig. 1g, Extended Data Fig. 2a; see Extended Data Fig. 2b–d for human primary bronchial HBEpC and nasal HNEpC epithelial cells, and the relationship between multiplicity of infection (MOI) and SA-β-gal). To closely mimic SARS-CoV-2 infection, we exposed ACE2-expressing HNEpC cells to SARS-CoV spike-protein-pseudotyped VSV glycoprotein G-deletion mutants (VSV-ΔG*/CoV-S and VSV-ΔG*/CoV-2-S). These pseudotypes all induced senescence in the cells in a spike-protein-dependent manner, as ACE2-negative NIH3T3 cells29 did not show senescence (Fig. 1h, Extended Data Fig. 2e). Retrovirus- and VSV-induced senescent cells exhibited increased levels of reactive oxygen species (ROS) and γ-H2AX DNA damage foci (Extended Data Fig. 2f). Conversely, the ROS scavenger N-acetylcysteine reduced γ-H2AX foci and abrogated VIS. Similarly, GS-441524, the active metabolite of the viral RNA polymerase inhibitor remdesivir, reduced γ-H2AX foci and blunted cGAS–STING pathway activation in VSV-infected cells (Extended Data Fig. 2g, h).
Importantly, HNEpC cells infected with the authentic SARS-CoV-2 virus also displayed SASP-positive senescence that was prevented by p53-shRNA-mediated inhibition of senescence or by GS-441524 treatment (Fig. 1i, j, Extended Data Fig. 2i, j). Moreover, RNA-seq analysis of SARS-CoV-2-infected alveolar epithelial cells (AECs) and publicly available RNA-seq datasets of normal human bronchus epithelial (NHBE) cells and lung cancer cell lines4 demonstrated that they also show robust signs of senescence induction (Fig. 1k, Extended Data Fig. 2k). Thus, virus infection in general and SARS-CoV-2 infection of respiratory epithelial cells in particular induce full-featured cellular senescence, which underscores our hypothesis that VIS might play a critical pathogenic part in COVID-19 lung disease.
Cellular senescence in patients with COVID-19
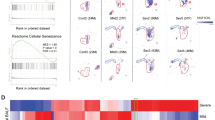
Next, we asked whether VIS would be a typical feature detectable in patients infected with SARS-CoV-2. We specifically analysed nasopharyngeal and lung tissue specimens from patients with a fatal course of PCR-proven COVID-19. Immunohistochemical analysis (IHA) of the senescence markers lipofuscin, p16INK4A, p21CIP1 and H3K9me3, and interleukin-8 (IL-8; encoded by CXCL8) as a representative SASP marker, showed robust reactivity in samples from infected individuals with or without remaining SARS-CoV-2 RNA in situ but not in individuals without COVID-19 (Fig. 2a, Extended Data Fig. 3a). Single-cell RNA-seq (scRNA-seq) analyses of upper airway mucosa samples provided further evidence of a senescence switch in patients with COVID-19 (ref. 30). The nasopharyngeal specimens showed increased transcript levels of p16INK4A (encoded by CDKN2A) and multiple SASP factors not only in SARS-CoV-2-permissive ciliated respiratory epithelial cells but also in macrophages, which implies that paracrine senescence spread to SASP-attracted macrophages and to other less virus-susceptible p16INK4A-elevated mucosal epithelial cell types (Fig. 2b). Gene expression profiling (GEP) of upper and lower airway specimens showed SASP-reminiscent cytokines in COVID-19 specimens induced at the transcript level (Fig. 2c). scRNA-seq data further showed that predominantly ciliated epithelial cells and macrophages scored positive for p16INK4A transcripts (Extended Data Fig. 3b).
a, IHA of senescence markers and BCL-w, and in situ hybridization of SARS-CoV-2 RNA in the respiratory mucosa of nasopharyngeal specimens from patients with COVID-19 (n = 6 for p16INK4A and IL-8, otherwise n = 5) compared with individuals without COVID-19 (non-COVID-19; n = 2). Quantification of affected areas is presented as the mean percentage of positive cells ± s.d. or the mean percentage of positive area ± s.d. (lipofuscin only). b, scRNA-seq-based GEP of upper airway tissue from patients with COVID-19 (n = 19) and healthy donors (n = 5). Colour-coded average-scaled (z scores) expression levels of the respective transcripts and cell types are shown. nrMa, non-residential macrophage; rMA, residential macrophage. c, GEP of SASP-reminiscent cytokines in upper (nasopharynx) and lower airway (lung) tissue from autopsy samples from patients with COVID-19 compared with individuals without COVID-19 (n = 3 each) by oligonucleotide-based transcript hybridization. d, CD86 IHA of macrophages in lung specimens from patients with COVID-19 (n = 13) compared with individuals without COVID-19 (n = 6). Mean number of positive non-epithelial cells in 0.1 mm2 ± s.d., P = 0.0005. e, Multiplex bead-based protein analysis of serum samples from patients with COVID-19 (n = 32) and healthy donors (n = 8). f, Mass spectrometry-based proteomics analysis of serum from patients with COVID-19 (n = 26) and from healthy donors (serum n = 3 and plasma n = 11). Heatmaps in e and f present scaled (z scores) protein intensities. g, Three-dimensional plot showing serum levels of D-dimer (clotting activity), SERPINA3 (coagulation-related SASP factor) and IL-6 (pro-inflammatory SASP factor) correlated to clinical COVID-19 severity per World Health Organization grades (g3–g6). n = 133 patients. The size of the sphere is proportional to the population size. Scale bars, 50 μm (a, d).
Key components of severe COVID-19 are a cytokine storm and macrophage activation syndrome3. We reasoned that the SASP of senescent upper airway epithelial cells may attract and activate macrophages through paracrine senescence induction31. This then contributes to the escalation of macrophage activation syndrome, a condition in which macrophages subsequently infiltrate the lungs. Notably, lungs of patients with COVID-19 showed infiltration by pro-inflammatory CD86+ macrophages and an M1-macrophage polarization profile32,33 (Fig. 2d, Extended Data Fig. 3c). In serum samples taken from patients with COVID-19, SASP peptides such as IL-8, PAI-1 (also known as serpin E1), CCL2 (also known as MCP-1), MMP-1, MMP-9 and TIMP-1 were among the most strongly upregulated factors (Fig. 2e). Selective scanning for clotting-relevant factors by ultrahigh-throughput mass-spectrometry-based proteomics34 also showed that these factors were induced to a large degree in COVID-19 serum probes (Fig. 2f). This result is in line with the transcriptional skewing of the Gene Ontology terms ‘coagulation’ and ‘complement’ in SARS-CoV-2-infected NHBE cells and in retrovirally senescent and even OIS or irradiation-senescent HDF cells7 (Extended Data Figs. 1b, 2k). Further underscoring the pro-thrombogenic propensity in patients with COVID-19, we detected elevated CD62+-marked platelet activation in their blood samples (Extended Data Fig. 3d). Accordingly, clinically more severe cases of COVID-19 exhibited a tight association of high SASP serum levels, clotting-relevant factors and activated coagulation35 (Fig. 2g, Extended Data Fig. 3e, f). There was also a close correlation between senescence marker transcripts and microthrombosis in lungs affected by COVID-19 in situ (Extended Data Fig. 3g, h). Thus, the findings from samples of patients with COVID-19 revealed alterations consistent with VIS and its SASP in the upper airway epithelia. Moreover, the results suggest that SASP-mediated effects are critical contributors to the respective lung disease and associated pro-inflammatory, tissue-damaging and pro-thrombogenic features.
Linking COVID-19 features to VIS
Next, we mechanistically interrogated whether supernatant from VIS cells (SNVIS) might induce COVID-19-typical functionalities in critical target cell types. Indeed, human THP-1 macrophages turned into a CD86+ or CD13+ M1-like state32 when incubated with SNVIS, but not supernatant from non-infected proliferating cells (SNprol.) or genetically senescence-incapable virus-infected HDFs (Fig. 3a, Extended Data Fig. 4a). The HDFs exhibited a strong SASP-reminiscent secretory profile, which is indicative of paracrine senescence, as only replication-incompetent viruses were used here (Fig. 3b, Extended Data Fig. 4b). Notably, transcripts reminiscent of SNVIS-induced CD86+-associated THP1 were similarly detected in lungs of patients with COVID-19 (Fig. 3c). Indeed, SNVIS exposure enforced paracrine, SA-β-gal-positive senescence in macrophages (Fig. 3d), which is all in line with their contribution as a VIS-triggered SASP-amplifying cellular messenger in COVID-19. Underscoring the critical role of an altered endothelium as a cytokine-releasing relay36, SNVIS also induced paracrine senescence in HUVEC human endothelial cells, which—in turn—launched a pro-inflammatory and pro-thrombogenic SASP (Fig. 3e, Extended Data Fig. 4c). Consistent with the microangiopathic thrombosis observed in severely affected lungs of patients with COVID-19, transfer of SNVIS, but not SNprol. or supernatant from virus-infected senescence-incapable HDFs, prompted platelet activation37 (Fig. 3f), accelerated clotting time (Fig. 3g) and provoked NET formation in vitro9 (Fig. 3h). Moreover, we observed enhanced formation of the C5b–C9 membrane attack complex and its ultimate cytotoxic action on HUVEC cells in response to SNVIS-activated human serum (Fig. 3i, j). This result mirrored the strongly elevated complement-activating factors in samples from patients with COVID-19. In essence, the pathogenesis of COVID-19 lung disease appears to be closely linked to senescence-governed immune escalation and pro-thrombotic effects.
a, The percentage of CD86+ cells in the THP-1 macrophage population exposed to supernatant from retroviral VIS compared with proliferating (Prol.) HDF IMR90 (as in Fig. 1a) and WI38 (as in Extended Data Fig. 1a) (left). Same experiment but using the supernatant of senescence-incapable IMR90 cells after mock or virus infection, as in Fig. 1e (right). b, Multiplex bead-based protein analysis of the supernatant of THP-1 exposed to supernatant as in a. SNVIS without THP-1 cells is shown on the right. c, GEP of M1-macrophage polarization-related transcripts by RNA-seq of THP-1 cells exposed to SNprol. or SNVIS and sorted for CD86 positivity (n = 2 each, left) and by oligonucleotide-based transcript hybridization of lung tissue from autopsy samples of patients with COVID-19 compared with individuals without COVID-19 (n = 3 each, right). d, SA-β-gal staining (top) and the CD86+ fraction (bottom) of THP-1 cells exposed to the supernatant of RPE1-infected VSV-ΔG* variants as indicated. e, SA-β-gal staining (top) and transcript expression by RT–qPCR (bottom) of HUVEC endothelial cells incubated with the supernatant of IMR90 cells. f, Platelet activation measured by ADP release from healthy donor platelets incubated with the supernatant of IMR90 cells as in a. n = 5 donors. g, Clotting time in whole-blood samples (n = 4 healthy donors) mixed with the indicated supernatant volumes (WI38, as in b). INTEM captures intrinsic, platelet-dependent clotting, whereas EXTEM captures extrinsic, non-platelet-dependent clotting. h, NET formation after exposing neutrophils to supernatant from HDFs as in b. n = 4 healthy donors, two measurements each. MPO, myeloperoxidase. i, Quantification of the cytolytic complement complex C5b–C9 on HUVEC cells incubated with human serum and supernatant as indicated. ina, inactivated. j, Cytotoxicity assay of HUVEC cells as in i. All quantifications (mean ± s.d. with individual values as dots) and representative photomicrographs are from n = 3 independent experiments unless otherwise noted. *P < 0.05 by unpaired two-tailed t-test (a, f, h–j), unpaired one-tailed t-test (d) or paired two-tailed t-test (g). Scale bars, 100 µm.
Eliminating VIS cells using senolytic drugs
On the basis of these findings, we considered VIS cells as a central therapeutic target in SARS-CoV-2 infection, and the early elimination of these cells might mitigate the course of disease. Previously, we and others presented evidence that senolysis (that is, the selective depletion of senescent cells) by drugs such as bafilomycin, ABT-263 (a BCL-2 family inhibitor; also known as navitoclax) or fisetin and quercetin (flavonoids that inhibit multiple kinases, with the latter typically in combination with the tyrosine kinase inhibitor dasatinib), reduced undesirable local and system-wide effects that persistent senescent cells might otherwise exert38,39,40,41,42. We first profiled transcript and protein expression of anti-apoptotic BCL-2 family members in VIS cells and found that BCL-xL (encoded by BCL2L1), BCL-w (encoded by BCL2L2) and MCL-1 (encoded by BCL2L3) were induced (Fig. 4a, Extended Data Fig. 5a; see also Fig. 2a, Extended Data Fig. 3a, b for elevated BCL2L1 and BCL2L2 expression in patients with COVID-19). This result suggests that navitoclax can selectively kill VIS cells. We also found that the PI3K–AKT–p70S6K axis, p38MAPK and the tyrosine kinase SRC were deregulated in VIS; therefore these serve as potential targets, especially for fisetin and for the combination of quercetin plus dasatinib (D/Q), according to predicted protein–chemical interactions43,44,45 (Fig. 4a, Extended Data Fig. 5b). Indeed, navitoclax, fisetin and D/Q were selectively cytotoxic in retrovirus-driven and VSV-driven VIS cells, but not in equally infected senescence-incapable HDFs in vitro (Fig. 4b, Extended Data Fig. 5c–e). By contrast, the BCL-2-specific venetoclax (also known as ABT-199) did not have these effects in the VIS cells. After senolysis, the release of infectious virions is a potential concern; however, we did not find enhanced infectivity after lysis of VIS cells (Extended Data Fig. 5f, g).
a, Immunoblots of BCL-2 family members (left) and kinases (right) in HDF lysates as indicated (α-tubulin was used as a loading control). b, The relative viability of IMR90 cells, as in Fig. 1e, 48 h after treatment with senolytics as indicated or with solvent-only (dimethyl sulfoxide (DMSO)). c, Viability of HNEpC-hACE2 cells infected with SARS-CoV-2 or mock, and treated with senolytics as indicated for c–f. d, The relative change of the CD86+ fraction in the THP-1 cell population after receiving supernatant from HNEpC-hACE2 cells as in c. Of note, supernatant transfer experiments were carried out in the presence of SARS-CoV-2-neutralizing anti-spike antibodies, which prevented the transmission of infection (as evidenced by negative target cell SARS-CoV-2 PCR, data not shown). e, Quantification of SA-β-gal staining of HUVEC endothelial cells after receiving supernatant from HNEpC-hACE2 cells as in d. f, Quantification of C5b–C9 on HUVEC cells exposed to human serum with supernatant from HNEpC-hACE2 cells as in d. All bar plots in this figure show the mean ± s.d. of n = 3 independent experiments with individual values as dots. *P < 0.05 by unpaired two-tailed t-test.
Importantly, infection by the authentic SARS-CoV-2 virus rendered HNEpC cells susceptible to navitoclax, fisetin and D/Q (Fig. 4c). Moreover, as seen above with other viruses, supernatant transfer from SARS-CoV-2-infected cells expanded CD86+ THP-1 macrophages and led to paracrine senescence or complement-mediated lysis of HUVEC cells. These effects were not observed when supernatants of epithelial cells pretreated with p53 shRNA or GS-441524 or exposed to any of the three senolytic treatment options were used for transfer (Fig. 4d–f, Extended Data Fig. 5h; see Extended Data Fig. 5i–l for VIS, SASP and senolytic susceptibility in the context of the SARS-CoV-2 variants Alpha and Beta, which have mutations that affect the spike protein).
Senolytic targeting of COVID-19 in vivo
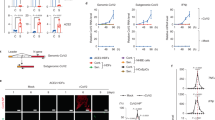
To determine the impact of senolytic treatment on the clinical severity of COVID-19-like lung disease, we first tested early intervention with navitoclax in the well-established Syrian golden hamster model of SARS-CoV-2-driven lung disease46,47. After intranasal infection with SARS-CoV-2, animals developed COVID-19-like pneumonia within the first 3–5 days. scRNA-seq profiling of SARS-CoV-2-infected and mock-infected hamster lungs revealed upregulated p16INK4A, core senescence and SASP factors, as well as Src transcript expression in alveolar epithelial cells, macrophages and neutrophils due to primary virus infection or paracrine senescence induction during days 3–5 after infection (Extended Data Fig. 6a). This analysis also demonstrated induction of BCL2L2 transcripts in these cell types at day 5 after infection (Extended Data Fig. 6b), which provides support for the use of navitoclax as a suitable senolytic. Animals on the navitoclax protocol exhibited moderate adverse effects (Extended Data Fig. 6c, d). However, they had a marked reduction in the number of senescent cells and improvements in the histopathological features compared with the solvent-only-treated but SARS-CoV-2-infected cohort on day 7 after infection (Fig. 5a, Extended Data Fig. 6e–g, Extended Data Table 1). We also detected much lower concentrations of key pro-inflammatory SASP factors in the serum of navitoclax-treated hamsters on day 7 after infection, values that were almost indistinguishable from uninfected controls (Extended Data Fig. 6h).
a, H3K9me3 IHA in nasopharyngeal (top) and lung (middle) specimens, and lipofuscin-based detection (bottom) of senescent cells in the lungs of golden hamsters infected with SARS-CoV-2 and treated with navitoclax or solvent-only (n = 5 animals each), or mock-infected and untreated (n = 3 animals) at day 7 after infection. b, H3K9me3 IHA and lipofuscin staining in nasopharyngeal sections of Roborovski dwarf hamsters infected with SARS-CoV-2 or mock at day 4 after infection and treatment as indicated. c, Lung tissue of dwarf hamsters as in b on day 4 after infection stained with haematoxylin and eosin. Uninfected healthy hamsters (mock) are shown for comparison ((xiv)–(xvii)). (i), (v) and (x) are overviews of lung sections. Signs of COVID-19-like pneumonia and haemorrhage are indicated: hyperplasia of the bronchial epithelium (double-headed arrows), haemorrhage (asterisks) and necrosis (black arrowheads) of alveolar epithelial cells with neutrophil infiltration (white arrowheads), and endothelialitis (black arrows). d, Quantification of the findings in c. *P < 0.05 by unpaired t-test (one-tailed). e, Multiplex bead-based protein analysis of dwarf hamster serum samples on day 4 after infection (as in b–d). n = 5 each (b–d; n = 4 each in e) for SARS-CoV-2-infected and treated (with solvent (Sol.) or navitoclax (Navito.) animals. n = 2 for healthy (mock) control. f, Constellation plots of patients with COVID-19 (ref. 52) before (day 0) and after 7 days of standard care (SC) only or quercetin plus SC. Variables (Extended Data Table 2) were simultaneously analysed by agglomerative hierarchical clustering to visualize similarity (distance) between patients and separation of the cohorts after treatment. Multi-dimensional analysis of individual patient data, with each red or green dot indicating a patient, the line length represents the distance between patients, and axes showing arbitrary units. n = 21 for each group. All quantifications are mean ± s.d. Scale bars, 20 µm (c, middle two columns), 50 µm (a, b, c (right column)) or 1 mm (c, left column).
To evaluate the potential of senolytic intervention in model systems with a more severe COVID-19-like clinical course, we next considered the Roborovski dwarf hamster model48. On day 4 after infection and treatment with navitoclax, D/Q or solvent-only, animals with senolytic intervention presented with a substantial reduction of H3K9me3- or lipofuscin-marked senescent cells in their respiratory epithelium and a profound reduction in lung disease features, especially in the D/Q-treated group (Fig. 5b–d). Both senolytic regimens also produced a reduction in SASP-reminiscent serum cytokines on day 4 after infection in this model, to levels close to healthy controls (Fig. 5e). Effects in the D/Q-treated group were particularly meaningful: 5 out of 5 animals survived, displayed no apparent signs of disease and no or only moderate weight loss (Extended Data Fig. 7a). By contrast, 4 out of 5 animals in the navitoclax-treated group and 3 out of 5 animals in the placebo-treated group had to be euthanized due to weight loss and clinical impairment by day 4 after infection. Last, we investigated the K18-hACE2-transgenic mouse model, in which SARS-CoV-2 infection causes severe and typically lethal disease49,50. Mice were treated in a similar manner to the dwarf hamster model, with oral administrations of navitoclax, D/Q or fisetin, and scheduled for lung analysis on day 6 after infection. For the solvent-only-treated mice, 2 out of 5 died on or before day 6, whereas 1 out of 5 of the navitoclax-treated mice died and the weight of another fell underneath the accepted weight limit on day 6. By contrast, all the D/Q-treated and fisetin-treated mice were alive, had limited weight loss and presented without clinical symptoms on day 6 (Extended Data Fig. 7a, b). Although overall histopathological analyses did not show overt differences regarding COVID-19-like parameters (Extended Data Fig. 7c; see Extended Data Fig. 7d regarding virus loads in this model and the hamster models), markers of senescent cells were significantly lower in the lungs of senolytic-treated mice across all treatment regimens (Extended Data Fig. 7e).
Quercetin was recently tested as an oral formulation with sunflower lecithin against standard care in two randomized clinical trials (ClinicalTrials.gov identifiers NCT04578158 and NCT04861298) for patients with confirmed SARS-CoV-2 infection and mild COVID-19-associated symptoms. In both trials, based on 152 patients51 and 42 patients52, senolytic intervention led to clinical improvement51,52. Agglomerative hierarchical clustering of data from the second trial52 across numerous parameters separated the patient population—which was indistinguishable at baseline—into quercetin and standard care at day 7 (Fig. 5f, Extended Data Table 2). Moreover, collective analysis of all 194 patients concluded that the quercetin-treated group showed significant risk reductions regarding the needs of hospitalization and oxygen therapy compared to standard care, particularly in terms of length of hospitalization, referral to the intensive care unit and number of deaths (Extended Data Fig. 8a, b). Together, early senolytic intervention during SARS-CoV-2 infection significantly attenuates COVID-19-like lung disease and systemic inflammation.
Our investigation marks VIS as a central pathogenic principle and a valid therapeutic target for SARS-CoV-2 infection to prevent severe COVID-19 lung disease. The primary virus–host cell encounter induces a senescence response associated with massive pro-inflammatory cytokine secretion that complements virus spreading to the lower airway tract. Our data pinpoint macrophages as pivotal cellular messengers in this process. Attracted to and primed by upper airway VIS cells, macrophages undergo a secondary, SASP-amplifying senescent-state switch. Subsequently deployed in the lungs, they further induce senescence, particularly in endothelial cells. System-wide and locally active pro-coagulatory and complement-activating SASP factors contribute to hallmark histopathological features and the clinical severity of COVID-19. Hence, early senolytic intervention is an appealing strategy to interrupt such an escalating immune cascade at its beginnings. The effects of D/Q, fisetin and navitoclax treatment that we observed here are encouraging, but optimized dose scheduling to maximize their clinical potential and limit potential toxicities is needed. Owing to their good safety profiles, compounds such as the flavonoids quercetin and fisetin are particularly attractive candidates as VIS-targeting senolytics. As already demonstrated for quercetin51,52, fisetin is currently being investigated in COVID-19 trials. Robbins and colleagues53 recently reported enhanced SASP production of pre-existing senescent cells in old mice by pathogen-associated molecular pattern factors such as lipopolysaccharide or the SARS-CoV-2 spike protein. In turn, pharmacological removal of senescent cells by fisetin or D/Q in β-coronavirus-infected mice was clinically effective. This adds to the rationale of senolytic VIS cell elimination, as proposed here, for SARS-CoV-2-induced senescence. Moreover, it will be interesting to explore whether early senolytic intervention can mitigate chronic post-COVID-19 impairment, which is known as long COVID54. Finally, we highlight that although effective vaccinations are available worldwide and will hopefully make COVID-19 history soon, our approach remains conceptually valid in the context of other viral epidemics or pandemics in the future.
Methods
Cell culture and virus production
Cell lines were maintained according to the guideline of the suppliers. The HDF cell lines IMR90, WI38 and Tig3 have been previously described22. The human cell lines A549 and HUVEC were purchased from the American Type Culture Collection (ATCC) and RPE1 (hTERT RPE1) was provided by A. Schiebel. Primary human nasal or bronchial epithelial cells (HNEpC and HBEpC) were purchased from PromoCell. HNEpC cells with exogenous ACE2 expression (HNEpC-hACE2) were generated by lentiviral infection (hACE2 lentivirus, BPS Bioscience). THP-1 cells were provided by A. Walter. Senescence-incapable cell lines were generated by transduction with lentiviruses encoding JMJD2C or p53 shRNA as previously described22,55.
All viruses used in this work are tagged with green fluorescent protein (GFP), with the exception of HCoV-NL63 and SARS-CoV-2, and infection efficiency was monitored by detecting fluorescence levels in infected cells. Retrovirus was produced by transfecting Phoenix packaging cells with a murine stem cell virus (MSCV) GFP plasmid56. Lentivirus was produced by transfecting 293T cells with psPAX2, pMD2.G (Addgene, 12260 and 12259) and pCDH-CMV-MCS-EF1-GFP plasmids22. AAV was obtained from the Charité Viral Core Facility. Caco-2 cell-adapted wild-type HCoV-NL63 virus at a titre of 3 × 106 PFU ml–1 was provided by the M.L. laboratory. VSV-GFP was a gift from W. Brune. VSV-ΔG* GFP (Kerafast) and plasmids encoding SARS-CoV spike (pKS SARS St19; provided by S. Fukushi), SARS-CoV-2 spike (Wuhan SARS-CoV-2 spike deltaCT28; provided by C. Ratswohl and K. de la Rosa), SARS-CoV-2 spike variants Alpha and Beta (pCDH-CMV-spike-B117-EF1-puro and pCDH-CMV-spike-B1351-EF1-puro synthesized by GENEWIZ) proteins were used to generate mutant pseudotypes. SARS-CoV-2 (hCoV-19/Germany/BY-ChVir-984/2020, sequence reference EPI_ISL_406862) for in vitro applications was propagated in Vero E6 cells (ATCC, CRL-1586) and genome-sequenced to exclude stocks with mutations from further experiments. Infection was monitored by PCR, to detect viral genomic RNA in the supernatant 48 or 72 h after infection, as previously described57. Experiments were conducted under appropriate biosafety conditions with enhanced respiratory personal protection equipment in the BSL-3 facility at the Institute for Virology, Charité, Berlin. In-house generated virus stocks were concentrated ten times using virus concentration kits (TaKaRa Bio and Abcam). High-titre virus infection to induce VIS reflect MOI of 50 for replication-incompetent retrovirus and lentivirus, 10 for replication-incompetent AAV and VSV-ΔG*, 10 for replication-competent HCoV-NL63, 1 for replication-competent VSV, and 0.1 for replication-competent SARS-CoV-2. VIS features were assessed on day 5 (retrovirus and lentivirus), day 3 (HCoV-NL63, VSV-ΔG* and SARS-CoV-2) or day 2 (VSV) after infection.
Patients with COVID-19, individuals without COVID-19 and healthy donor material
Upper (nasopharyngeal) and lower airway (lung) tissue formalin-fixed paraffin-embedded (FFPE) samples were derived from deceased patients infected with SARS-CoV-2 and from archive material from patients who passed away without notable clinical signs of a respiratory infection before October 2019 (that is, individuals without COVID-19), blood samples of patients with SARS-CoV-2 and hospitalized patients in the course of their COVID-19 disease, or blood samples from healthy volunteers were used based on approval by the local ethics commission of the Johannes Kepler University (reference number 1070/2020). All patients with COVID-19 were PCR-proven as SARS-CoV-2-positive; however, SARS-CoV-2 RNA was detectable in 4 out of 5 cases of COVID-19 shown in Fig. 2a. All participants gave informed consent. In addition, we conducted re-analyses of additional patient and control materials that were previously obtained and published elsewhere30,34,51,52.
Flow cytometry, immunoblotting, immunofluorescence and immunohistochemical analysis
Flow cytometry, immunoblotting (IB), immunofluorescence (IF) and IHA were performed as previously described24,58. Surface antigen detection by flow cytometry was carried out using fluorescence-conjugated antibodies against human CD86-PE-Cy7 (BioLegend, 374209; 1:200), CD13-APC-Cy7 (BioLegend, 301709; 1:200) and CD62P-PE (BioLegend, 304905; 1:200). Antigen detection by IB was carried out using the following antibodies: BCL-2 (Cell Signaling Technology (CST), 15071; 1:1,000); BCL-xL (CST, 2764, 1:1,000); BCL-w (CST, 2724; 1:1;000); MCL-1 (CST, 39224; 1:1,000); phospho-p65-Ser536 (CST, 3033; 1:1,000); phospho-IRF3-Ser386 (CST, 37829; 1:1,000); phospho-Rb-Ser807/811 (CST, 8516; 1:1,000); p21CIP1 (Santa Cruz Biotechnology (SCBT), sc-6246; 1:200); p16INK4A (SCBT, sc-1661; 1:200); H3K9me3 (Abcam, ab8898; 1:2,000); phospho-Akt (Thr308) (CST, 13038T; 1:2,000); phospho-p38 MAPK (Thr180/Tyr182) (CST, 4511T; 1:1,000); phospho-p70 S6 kinase (Thr389) (CST, 9234T; 1:1,000); phospho-SRC (Tyr416) (CST, 6943T; 1:1,000); and α-tubulin (Sigma, T5168; 1:2,000). Anti-mouse or anti-rabbit horseradish-peroxidase-conjugated antibodies were used as secondary antibodies (GE Healthcare, RPN4301 and NA931V, respectively). Antigen detection by IF was performed using the following antibodies: phospho-H2A.X (CST, 9718; 1:400); p16INK4A (Abcam, ab211542; 1:250); C5b–C9 (SCBT, sc-58935; 1:250); anti-mouse-Cy3 secondary antibody (Thermo Fisher, A10521; 1:1,000); and anti-rabbit-594 secondary antibody (Thermo Fisher, A11012; 1:1,000). Antigen detection by IHA was performed using a HiDef Detection HRP Polymer System (Cell Marque, 954D-30) with antibodies against p21CIP1 (SCBT, sc-6246; 1:200), IL-8 (Abcam, ab18672; 1:200), H3K9me3 (Abcam, ab8898; 1:2,000), BCL2L2 (Abcam, ab190952; 1:1,000) and CD86 (Antibodies-online, ABIN736701; 1:100).
Original scans of the immunoblots presented in Figs. 1f, 4a, with molecular mass markers and crop area indicated, are shown in the Supplementary Information.
In situ hybridization
In situ hybridization was performed as previously described46 using a ViewRNA Tissue Assay Core kit (Invitrogen, 19931) and a ViewRNA Tissue Assay Blue Module (Invitrogen, 19932) according to the manufacturer’s instructions. For the detection of SARS-CoV-2, ViewRNA Type 1 Probe Set COVID19 polyprotein (Invitrogen, CVX-06, Assay ID VPNKRHH) and ViewRNA Type 6 Probe Set Human ACTB (Invitrogen, VX-06, Assay ID VA6-10506) were used. Following hybridization, sections were counterstained with haematoxylin and mounted with ProLong Glass Antifade Mountant (Invitrogen, P36982). Images were taken using an Olympus Slide Scanner VS200.
Quantification of gene transcription
RT–qPCR
RNA isolated from cell pellets using TRIzol (Invitrogen) was transcribed to cDNA using SuperScript II reverse transcriptase (Invitrogen). Individual gene expression was analysed by RT–qPCR using Taqman assays from Applied Biosystems. A list of the Taqman assays used is available in the Supplementary Information.
RNA-seq
Total RNA was isolated using a RNAeasy Mini kit (Qiagen) and sequenced at BGI Genomics. scRNA-seq was conducted as previously described59. Further processing of the raw sequencing data is described in the section ‘Statistics and data analysis’.
Oligonucleotide-hybridization-based transcriptome profiling
RNA was extracted from FFPE tissue sections using a PureLink FFPE RNA Isolation kit (Thermo Fisher Scientific, K156002). RNA was then hybridized with the probe set of the NanoString nCounter Human Tumor Signaling 360 Panel (XT-CSPS-H-T360-12) and analysed on a NanoString nCounter SPRINT.
Quantification of protein expression
Multiplex bead-based protein detection (MAGPIX)
MAGPIX assays were conducted according to the manufacturer’s manual with following bead panels: custom Procartaplex 19-plex (Thermo Fisher Scientific, PPX-19, Assay ID MXAACDR), custom Procartaplex 4-plex (Thermo Fisher Scientific, PPX-04, Assay ID MXCE4XN) and Bio-Plex Pro Rat Cytokine 23-Plex Assay (Bio-Rad, 12005641). The assay was measured using a BioPlex MAGPIX multiplex reader (Bio-Rad) and analysed with a 5PL algorithm provided by the Bio-Plex Manager software (v.6.1). Detailed procedures for sample preparation and measurement are described in the Supplementary Information. Note that owing to a different normalization procedure, values cannot be cross-read between different panels (for example, Fig. 1d compared with Fig. 3b).
Mass-spectrometry-based proteomics
Detailed protocols of sample preparation, mass spectrometry and data processing have been previously described34; for further details see the Supplementary Information. In brief, serum samples were reduced, alkylated, digested and conditioned on a Biomek i7 workstation, and approximately 5 µg of peptides per injection was used for the gradient liquid chromatography–tandem mass spectrometry analysis. The raw data were processed using DIA-NN 1.7.12.
VIS assays in vitro
Growth curve analysis
The HDF IMR90, WI38 or Tig3 cells were infected with MSCV retrovirus at day 0 and day 3, and SA-β-gal activity was measured at day 5. For growth curve analyses, 2 × 104 cells were seeded into 12-well plates and cell numbers were counted at days 3, 6 and 9.
2′3′-cGAMP ELISA assay
Cell cytoplasmic 2′3′-cGAMP concentration was measured using a Caymanchem ELISA kit. In brief, cells were lysed in M-PER extraction reagent (Thermo Fisher) and 10 μg protein lysate was used to determine the 2′3′-cGAMP content.
Analysis of cell viability and senescence
Viability and cell numbers were analysed by Trypan blue dye exclusion or Guava Viacount (Millipore). SA-β-gal activity at pH 6.0 (for human cells) or pH 5.5 (for rodent cells) as a senescence marker in fresh or cryopreserved cells was assessed as previously described60. In situ detection of lipofuscin as a senescence marker in FFPE samples was carried out using SenTraGor reagent (Lab Supplies Scientific; provided by V. G. Gorgoulis) according to the manufacturer’s instructions.
ROS detection
Dihydroethidium (20 μM, MedChemExpress) was directly added to cells and incubated at 37 °C for 20 min. Dihydroethidium was then washed away with PBS. Cells were fixed in 4% paraformaldehyde and staining intensity was analysed using a Nikon fluorescence microscope.
VIS supernatant transfer and secondary, paracrine senescence
Conditioned medium was collected from proliferating and VIS cells after 24 h of incubation and centrifuged at 5,000 r.p.m. for 5 min. The supernatant (SNprol. or SNVIS, respectively) was transferred to HUVEC or THP-1 cells for further analysis. For the supernatant transfer from SARS-CoV-2-infected HNEpC cells, custom-made neutralizing antibodies against SARS-CoV-2 (provided by the H.S. laboratory) were mixed in to block secondary infection.
NET formation assay
NET formation was measured according to the concentration of NET DNA bound to myeloperoxidase (MPO). Neutrophils were isolated from whole blood of four healthy donors. NET formation was quantified as previously described61. In brief, 10 µg ml–1 anti-MPO antibody (Abcam, ab25989) was immobilized on the surface of black MaxiSorp-treated plates (Thermo Scientific). The plate was subsequently washed with PBS and blocked with 5% non-fat milk for 2 h at room temperature. Neutrophils incubated for 4 h at 37 °C in RPMI medium from proliferating or VIS WI-38 or IMR-90 cells were pipetted onto the MPO-coated plate and incubated for 1 h at room temperature. NET formation quantification was performed using a MPO-DNA PicoGreen assay according to the manufacturer’s instructions (Quant-iT PicoGreen dsDNA Assay kit, Invitrogen). Sample fluorescence was measured using a GloMax Discover microplate reader (Promega).
Rotational thromboelastometry
To measure clotting time, a rotational thromboelastometry assay was performed on a ROTEM delta (Werfen) device according to the manufacturer’s instructions. INTEM captures intrinsic, platelet-dependent clotting and EXTEM extrinsic, non-platelet-dependent clotting. Whole-blood samples from healthy donors were spiked with the supernatant from WI38 cells in proliferation or VIS. Exclusion criteria for the healthy donors were coagulation disorders and clotting-affecting medication. Sex distribution was equal among the donors.
Platelet activation and ADP release measurement by VIS supernatant
Platelet activation was measured by flow cytometry as the percentage of CD62/P-selectin-positive cells in whole blood. To measure VIS-induced ADP release, 2 × 106 platelets from healthy donors were resuspended in 200 µl conditioned medium and incubated at 37 °C for 1 h. Supernatant was collected by centrifugation at 2,000g for 5 min, and the ADP concentration was measured using an ADP Colorimetric Assay Kit II (BioVision) according to the manufacturer’s instruction.
Complement-mediated cytotoxicity in HUVEC endothelial cells
Serum from healthy donors (500 µl) was incubated with proliferating or VIS HDF (Fig. 3i, j) or HNEpC cells (Fig. 4f, Extended Data Fig. 5h) in 5 ml medium at 37 °C for 30 min. Heat-inactivated (56 °C for 30 min) serum was used as a control. Supernatant was collected and incubated with HUVEC endothelial cells at 37 °C. After 2 h of incubation, HUVEC cells were fixed in 4% paraformaldehyde, and C5b–C9 immunofluorescence staining was carried out. Alternatively, 24 h after incubation, a cytotoxicity assay was performed using a CyQUANT LDH Cytotoxicity kit (Invitrogen).
Senolytic treatment in vitro and secondary viral infection
VIS cells were treated with venetoclax (1 µM), navitoclax (1 µM), fisetin (20 µM), D/Q (20 µM, reflecting 2 µM dasatinib plus 20 µM quercetin) or AT-101 (1 µM) for 48 h, and cell viability was determined by Trypan blue staining using a Countess II FL Automated Cell Counter (Thermo Fisher). To generate the supernatant used for secondary viral infection, wild-type mouse embryo fibroblasts were infected with VSV for 6 h and treated with 1 µM navitoclax for 24 h. Supernatant was centrifuged and filtered through a 0.22-µm PVDF filter and added to THP-1 cells. After 24 h, secondary infectivity was measured by counting GFP+ THP-1 cells.
SARS-CoV-2-driven COVID-19 animal models
In vitro and in vivo studies related to hamsters were conducted under appropriate biosafety conditions in the BSL-3 facility at the Institute for Virology, Freie Universität Berlin, Germany. All hamster experimentation was approved by the relevant state authority (Landesamt für Gesundheit und Soziales Berlin, Germany; permit number 0086/20), and performed in compliance with national and international guidelines for the care and humane use of animals. Preparation of SARS-CoV-2 virus stock and intranasal infection of Syrian hamsters (Mesocricetus auratus) as a faithful in vivo model of COVID-19 lung disease have been previously described46. In brief, a SARS-CoV-2 wild-type isolate (BetaCoV/Munich/BavPat2-ChVir984-ChVir1017/2020) was grown in Vero E6 cells. To ensure genetic integrity, passage 3 stocks were genome-sequenced, and results showed conformity with the published sequence (GenBank MT270112.1) and confirmed the presence of the furin cleavage site essential for in vivo pathogenesis. Anaesthetized male and female hamsters at 6–10 weeks of age were inoculated with 1 × 105 PFU SARS-CoV-2 in a total volume of 60 µl cell culture medium. Control animals were mock-infected with the same volume of cell culture medium. One day after infection, animals were randomized and half of the animals (n = 5) received a single 85 mg kg–1 body-weight dose of navitoclax in solvent (60% Phosal PG, 30% PEG 300, 20% ethanol) intraperitoneally every day for 6 consecutive days. The other half of infected animals (n = 5) received the same volume of solvent only. The control group (n = 3) did not receive virus or treatment. On day 7 after infection, 1 day after the last treatment, all hamsters were euthanized, and tissue and peripheral blood samples were prepared for further analyses.
The Roborovski dwarf hamster (Phodopus roborovskii), a species highly susceptible to a severe course of SARS-CoV-2 infection, was used under the permit mentioned above. Infections were performed as previously described48. Female Roborovski dwarf hamsters at 5–7 weeks of age underwent the procedure described above for Syrian hamsters with the exception that the total volume of the inoculum was reduced to 20 µl. The same virus stock was used. Animals were randomly distributed into three treatment groups (n = 5) and one non-infected, non-treated group (n = 2). After 6 h of infection, animals were treated orally once a day with navitoclax (85 mg kg–1) or D/Q (5/50 mg kg–1) dissolved in 60% Phosal PG, 30% PEG 300, 20% ethanol and mixed 1:1 with strawberry syrup. The placebo-treatment group received the same solvent/syrup combination without drug. On day 4 after infection, animals were killed, and tissue and peripheral blood specimens were collected for downstream analyses.
Infection of K18-hACE2 mice (Jackson Laboratory strain 034860) with SARS-CoV-2 was performed according to approval by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai as previously reported62. In brief, 6-week-old female mice were infected with 1 × 104 PFU SARS-CoV-2 (USA-WA1/2020, BEI resources NR-52281) grown in Vero E6 cells and validated by genome sequencing, and randomly distributed into three treatment groups (n = 5 each) and one solvent-only control group (n = 5). From 1 day after infection, randomly chosen animals were orally treated with oral navitoclax (85 mg kg–1), D/Q (5/50 mg kg–1) or fisetin (100 mg kg–1) dissolved in 60% Phosal PG, 30% PEG 300, 20% ethanol every other day. On day 6 after infection (1 day after the last treatment), animals were killed, and tissue specimens were collected for histopathological analyses.
Hamster and mouse lung tissues were processed for histopathology as previously described47 and analysed by board-certified veterinary pathologists (blinded about the treatment) following suggested standard criteria63. Transmission electron microscopy was performed with trachea tissue from infected and treated Syrian golden hamsters. Fixation and further processing of the samples and image acquisition procedures are described in the Supplementary Information.
Statistics and data analysis
Based on previous experience22,24,41,47,50,53,54,57, sample sizes typically reflected three to five replicates. In some experiments, much higher numbers of individual primary material were used as biological replicates. All quantifications from staining reactions were carried out by an independent and blinded second examiner and reflect at least 3 samples with at least 100 events counted (typically in 3 different areas) each. For patients and for animal models, IHA quantification was carried out in SARS-CoV-2-affected areas. Regarding Fig. 5, Extended Data Figs. 6 and 7, quantification of the indicated features was based on the following numbers of individual photomicrographs from animals as indicated in the legends and text: Fig. 5a: nasopharynx H3K9me3 (n = 3 for mock, n = 6 for solvent and navitoclax), lung H3K9me3 and lipofuscin (n = 3 for mock, n = 5 for solvent and navitoclax); Fig. 5b: H3K9me3 (n = 5 for solvent and navitoclax, n = 6 for D/Q), lipofuscin (n = 5 for all three groups); Extended Data Fig. 7e: p16INK4A (n = 5 for solvent, navitoclax and D/Q, n = 4 for fisetin), H3K9me3 (n = 4 for solvent and navitoclax, n = 5 for fisetin and D/Q), lipofuscin (n = 5 for solvent and navitoclax, n = 4 for fisetin, n = 7 for D/Q).
Data analysis was performed with GraphPad Prism 9. For statistical comparisons, a t-test was applied. Unless otherwise stated, a P value of <0.05 was considered statistically significant. For multiple-testing corrections, the Benjamini–Hochberg method was used to control for the false discovery rate64. Bioinformatics analysis was performed in R, v.4.0.3, and Bioconductor 3.12 using various R packages as described in the Supplementary Information in greater detail. High-performance computation was performed in the Research/Clinic cluster of the Berlin Institute of Health. For further analyses of the clinical data from patients with COVID-19 (refs. 51,52) as presented in Fig. 5f and Extended Data Fig. 8a, b, JMP Pro 14 (SAS Institute) was used for agglomerative hierarchical clustering, GraphPad Prism 9 was used for Fisher’s exact test for P values, and a Haldane–Anscombe correction was applied to calculate relative risks. Lipofuscin (SenTraGor) staining was quantified with the pixel counting function of the ImageJ program (bundled with Java 1.8.0_172, NIH)65. No statistical methods were used to predetermine sample size.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The WI38 VIS- and OIS-derived raw RNA-seq data (Fig. 1b and Extended Data Fig. 1b) were deposited at the Gene Expression Omnibus (GEO) repository of the National Center for Biotechnology Information (NCBI) under accession number GSE165532. Gene expression data of human AECs infected with SARS-CoV-2 (Fig. 1k) are publicly available from at the GEO under the accession number GSE148729. Raw sequencing data of NHBE, Calu-3 and A549 cells infected with SARS-CoV-2 (Extended Data Fig. 2k) are publicly available at the GEO under the accession number GSE147507 (ref. 4). Seurat objects comprising scRNA-seq datasets of nasopharyngeal, protected specimen brush and bronchial lavage samples of 19 patients with COVID-19 and 5 healthy donors30, constituting a total of 36 samples that was analysed in this study (Fig. 2b), are available at FigShare (https://doi.org/10.6084/m9.figshare.12436517; covid_nbt_main.rds, covid_nct_loc.rds). The mass-spectrometry-based proteomics data of serum from patients with COVID-19 and healthy donors in Fig. 2f are described in ref. 34. Patient proteomics data (Fig. 2g, Extended Data Fig. 3e) are publicly available at https://www.who.int/teams/blueprint/covid-19, in which disease severity was defined as follows: g3, hospitalized, no oxygen therapy; g4, oxygen by mask or nasal prongs; g5, noninvasive ventilation or high-flow oxygen; g6, intubation and mechanical ventilation; g7, ventilation and additional organ support. scRNA-seq data of hamsters infected with SARS-CoV-2 (Extended Data Fig. 6a, b) are available at the GEO (GSE162208)59. All information related to the two randomized trials on early quercetin intervention in patients with COVID-19 (NCT04578158 and NCT04861298) can be found in the respective publications51,52 and in Extended Data Table 2.
References
Zhang, X. et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 583, 437–440 (2020).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).
Merad, M. & Martin, J. C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020).
Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010).
Lee, S. & Schmitt, C. A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 21, 94–101 (2019).
Wiley, C. D. et al. SILAC analysis reveals increased secretion of hemostasis-related factors by senescent cells. Cell Rep. 28, 3329–3337.e5 (2019).
Ackermann, M. et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 383, 120–128 (2020).
Middleton, E. A. et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 136, 1169–1179 (2020).
Berlin, D. A., Gulick, R. M. & Martinez, F. J. Severe Covid-19. N. Engl. J. Med. 383, 2451–2460 (2020).
Wolfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020).
Ju, B. et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020).
Horby, P. et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 384, 693–704 (2021).
Xu, X. et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl Acad. Sci. USA 117, 10970–10975 (2020).
Chuprin, A. et al. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev. 27, 2356–2366 (2013).
Martínez, I. et al. Induction of DNA double-strand breaks and cellular senescence by human respiratory syncytial virus. Virulence 7, 427–442 (2016).
Baz-Martinez, M. et al. Cell senescence is an antiviral defense mechanism. Sci. Rep. 6, 37007 (2016).
Hsieh, T. H. et al. Senescence in monocytes facilitates dengue virus infection by increasing infectivity. Front. Cell. Infect. Microbiol. 10, 375 (2020).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003).
Yu, Y. et al. Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell 33, 322–336 (2018).
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008).
Jing, H. et al. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 25, 2137–2146 (2011).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Buss, H. et al. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-kB at serine 536 is mediated by multiple protein kinases including IkB kinase (IKK)-α, IKKβ, IKKε, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279, 55633–55643 (2004).
Wang, P. et al. Expression cloning of functional receptor used by SARS coronavirus. Biochem. Biophys. Res. Commun. 315, 439–444 (2004).
Chua, R. L. et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38, 970–979 (2020).
Acosta, J. C. et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 15, 978–990 (2013).
Roussel, M. et al. Mass cytometry deep phenotyping of human mononuclear phagocytes and myeloid-derived suppressor cells from human blood and bone marrow. J. Leukoc. Biol. 102, 437–447 (2017).
Li, X. et al. The flavonoid quercetin ameliorates liver inflammation and fibrosis by regulating hepatic macrophages activation and polarization in mice. Front. Pharmacol. 9, 72 (2018).
Messner, C. B. et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 11, 11–24.e4 (2020).
Demichev, V. et al. A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 12, 780–794.e7 (2021).
Kang, S. et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc. Natl Acad. Sci. USA 117, 22351–22356 (2020).
Beigi, R., Kobatake, E., Aizawa, M. & Dubyak, G. R. Detection of local ATP release from activated platelets using cell surface-attached firefly luciferase. Am. J. Physiol. 276, C267–C278 (1999).
Zhu, Y. et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015).
Xu, M. et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 24, 1246–1256 (2018).
Dorr, J. R. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Kirkland, J. L. & Tchkonia, T. Senolytic drugs: from discovery to translation. J. Intern. Med. 288, 518–536 (2020).
Freund, A., Patil, C. K. & Campisi, J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30, 1536–1548 (2011).
Bent, E. H., Gilbert, L. A. & Hemann, M. T. A senescence secretory switch mediated by PI3K/AKT/mTOR activation controls chemoprotective endothelial secretory responses. Genes Dev. 30, 1811–1821 (2016).
Jung, S. H. et al. Integrin α6β4–Src–AKT signaling induces cellular senescence by counteracting apoptosis in irradiated tumor cells and tissues. Cell Death Differ. 26, 245–259 (2019).
Kreye, J. et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell 183, 1058–1069.e19 (2020).
Osterrieder, N. et al. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses 12, 779 (2020).
Trimpert, J. et al. The Roborovski dwarf hamster is a highly susceptible model for a rapid and fatal course of SARS-CoV-2 infection. Cell Rep. 33, 108488 (2020).
Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020).
White, K. M. et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 371, 926–931 (2021).
Di Pierro, F. et al. Possible therapeutic effects of adjuvant quercetin supplementation against early-stage COVID-19 infection: a prospective, randomized, controlled, and open-label study. Int. J. Gen. Med. 14, 2359–2366 (2021).
Di Pierro, F. et al. Potential clinical benefits of quercetin in the early stage of COVID-19: results of a second, pilot, randomized, controlled and open-label clinical trial. Int. J. Gen. Med. 14, 2807–2816 (2021).
Camell, C. D. et al. Senolytics reduce coronavirus-related mortality in old mice. Science 373, eabe4832 (2021).
Nalbandian, A. et al. Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021).
Milanovic, M. et al. Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100 (2018).
Schmitt, C. A., Rosenthal, C. T. & Lowe, S. W. Genetic analysis of chemoresistance in primary murine lymphomas. Nat. Med. 6, 1029–1035 (2000).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT–PCR. Euro Surveill. 25, 2000045 (2020).
Schmitt, C. A., McCurrach, M. E., de Stanchina, E., Wallace-Brodeur, R. R. & Lowe, S. W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13, 2670–2677 (1999).
Nouailles, G. et al. Longitudinal omics in Syrian hamsters integrated with human data unravel complexity of moderate immune responses to SARS-CoV-2. Preprint at https://doi.org/10.1101/2020.12.18.423524 (2020).
Reimann, M. et al. Tumor stroma-derived TGF-β limits Myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17, 262–272 (2010).
Veras, F. P. et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 217, e20201129 (2020).
Rathnasinghe, R. et al. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 9, 2433–2445 (2020).
Gruber, A. D. et al. Standardization of reporting criteria for lung pathology in SARS-CoV-2-infected hamsters: what matters? Am. J. Respir. Cell Mol. Biol. 63, 856–859 (2020).
Hochberg, Y. & Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818 (1990).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Fridman, A. L. & Tainsky, M. A. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene 27, 5975–5987 (2008).
Hari, P. et al. The innate immune sensor Toll-like receptor 2 controls the senescence-associated secretory phenotype. Sci. Adv. 5, eaaw0254 (2019).
Schleich, K. et al. H3K9me3-mediated epigenetic regulation of senescence in mice predicts outcome of lymphoma patients. Nat. Commun. 11, 3651 (2020).
Acknowledgements
We thank S. Fukushi and M. Saijo, C. Ratswohl and K. de la Rosa, V. G. Gorgoulis, K. Hoffman and W. Brune, E. Schiebel for materials; F. Walper for their support in the BSL-3 facility; F. Kurth, P. Tauber-Lau, V. Demichev and L. E. Sander on behalf of the Charité PA–COVID-19 study group as well as members of the Charité Core Facility High-throughput Proteomics for data and measurements; S. Jangra, R. Rathnasinghe and R. Albrecht for support in the BSL-3 facility and procedures, and R. Cadagan for technical assistance; M. Almeder for handling patient samples; M. Recchia for the statistical analysis of the data from clinical trials; and members of the collaborating laboratories for discussions and editorial advice. This work was supported by grants to C.A.S. from the Medical Faculty of the Johannes Kepler University, Linz, Austria, the Deutsche Krebshilfe (no. 7011377629), the Deutsche Forschungsgemeinschaft DFG (GO 2688/1-1 | SCHM 1633/11-1, SCHM 1633/9-1), and the Förderverein Hämatologie und internistische Onkologie (Tyle Private Foundation, Linz, Austria), and to S.L. and C.A.S. by the German BMBF e:Med programme project SeneSys (no. 031L0189A). J. Trimpert and A.D.G. were funded by the German Research Council (grant SFB-TR84 Z01b). This interdisciplinary work was further made possible by the Berlin School of Integrative Oncology (BSIO) graduate programme funded within the German Excellence Initiative (with D.B. as a member of this programme) and the German Cancer Consortium (GCC). This research was also partly funded by CRIP (Center for Research on Influenza Pathogenesis), a NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract no. HHSN272201400008C) and CRIPT (Center for Research on Influenza Pathogenesis and Transmission), a NIAID-funded Center of Excellence for Influenza Research and Response (CEIRR, contract no. 75N93021C00014), by DARPA grant HR0011-19-2-0020, by supplements to NIAID grant U19AI142733, U19AI135972 and DoD grant W81XWH-20-1-0270, by a Fast Grant of the Mercatus Center and by the support of the JPB Foundation, the Open Philanthropy Project (research grant 2020-215611 (5384)) and anonymous donors to A.G.-S. M. Rasler is partly funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001134), the UK Medical Research Council (FC001134) and the Wellcome Trust (FC001134).
Author information
Authors and Affiliations
Contributions
S.L., Y.Y., F.B., M. Mairhofer, D.B., S. Kaltenbrunner, A.L., M. Schönlein, M. Reimann and D.N.Y.F. outlined and performed the molecular genetic, biochemical and cell biological experiments in cell lines, primary human and hamster material. J. Trimpert, J.M.A. and N.O. conceptualized and conducted the hamster experiments. P.R.-P. carried out the bioinformatics analyses. E.W., M.L. and R.E. conceptualized and generated the scRNA-seq data. M.P., S. Kunz, B.P., R.M. and R.L. analysed the tissue specimens from patients with COVID-19 and healthy donors. L.K., M. Mairhofer and J. Tomasits conceptualized and conducted the clotting-related experiments. T.C.F., K.D., E.G.M. and A.D.G. analysed the histopathology of hamster and mouse tissues. M. Schotsaert, C.M.-R., G.S., M.U. and A.G.-S. conceptualized and conducted the mouse experiments. D.N. and C.D. enabled SARS-CoV-2 infections of human cells and contributed to the experimental design. R.G., H.J.F.S. and B.L. provided clinical insights and blood samples from patients with COVID-19. C.P. and A.H. processed blood samples and carried out SARS-CoV-2 PCR testing. M. Mülleder and M. Ralser conducted the proteome data analysis and interpretation. S. Kimeswenger and W.H. contributed conceptual input and logistical support regarding specimen collection and virus-based experiments. A.K. and F.D.P. designed and conducted clinical trials on quercetin in patients with COVID-19. H.S. provided neutralizing antibodies targeting the spike protein of the SARS-CoV-2 virus and gave experimental recommendations in this regard. S.L. and C.A.S. designed the project, supervised the data analysis and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.U. contributed to this Article as an employee of Mount Sinai and the views expressed do not necessarily represent the views of Regeneron Pharmaceuticals. F.D.P. is a member of the Scientific Board of Pharmextracta, the vendor of Quevir, a dietary supplement containing quercetin in a sunflower lecithin formulation. The laboratory of A.G.-S. has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, and Merck. A.G.-S. has consulting agreements with the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, and Pfizer. A.G.-S. is an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections owned by the Icahn School of Medicine at Mount Sinai, New York. C.A.S. received travel support, honoraria and consulting fees from Abbvie, AstraZeneca, Bayer, Bristol-Myers Squibb/Celgene, Gilead/Kite, Janssen-Cilag, MSD, Novartis, Octapharma, Pierre Fabre, Roche, Sanofi, Takeda and TissUse.
Additional information
Peer review information Nature thanks Jesus Gill, Stanley Perlman and David Sinclair for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Additional biological properties of retroviral VIS.
a, SA-β-gal staining, SAHF formation by DAPI, and p16INK4A staining of HDFs (Tig3 and WI38) five days after retroviral infection or mock control. Representative photomicrographs of n = 3 independent experiments. b, Gene set enrichment analysis (GSEA) of virus infection-relevant GO terms probing RNA-seq datasets of WI38 HDF in VIS and OIS. Positive normalized enrichment scores (NES) indicate enrichment in VIS (dark grey bars) or OIS (light grey bars) compared to proliferating counterparts (mock infection for VIS, or empty vector control for OIS). NES with FDR q ≤ 0.1 are considered statistically significant and presented (for individual q-values, see Supplementary Information); n = 3 biological replicates each. c, Gene expression analysis for core senescence and SASP genes by RT-qPCR in WI38 and Tig3 as in a. Mean relative transcript level compared to mock control + s.d. of n = 3 independent experiments are shown. d, Growth curve analyses of HDF infected with retrovirus at different MOI as indicated, showing that high-titer virus induced VIS, reflected by stable cell numbers over time, while lower virus titer remained compatible with exponential cell growth. n = 3 independent experiments are presented as mean cell numbers ± s.d. e, SA-β-gal staining (left) and gene expression analysis of the indicated transcripts by RT-qPCR (right) in wild-type (WT) or senescence-defective p53-/- MEF, five days after infection with high-titer retrovirus or mock as a control. Representative photomicrographs with fractions of SA-β-gal-positive cells, and mean relative transcript levels normalized to mock control ± s.d. of n = 4 independent experiments are shown. f, Gene expression analysis of the indicated transcripts by RT-qPCR in IMR90 cells expressing JMJD2C, p53 shRNA (shp53), or control vector as in Fig. 1e. g, Multiplex bead-based protein analysis of SN of senescence-incapable IMR90 as in Fig. 1e. SNmock = SN of mock-infected cells; SNvirus = SN of retrovirus-infected cells. Mean expression levels of n = 3 biological replicates are shown. h, cGAS/STING activation upon viral infection, as evidenced by a higher induction of cyclic guanosine monophosphate-adenosine monophosphate (2'3'-cGAMP) by ELISA analysis in matched pairs of HDF after either mock or retrovirus infection (mean of n = 4 independent experiments for each cell line, upper panel). Mean cGAMP levels + s.d. for senescence-incapable IMR90 as in Fig. 1e (n = 4 independent experiments, lower panel). i, SA-β-gal staining of IMR90 cells, treated with reverse transcriptase inhibitor azidothymidine Zidovudine (50 µM), cGAS inhibitor G150 (5 µM), STING inhibitor H-151 (1 µM), or DMSO. Mock infection and DMSO solvent treatment as negative controls. Representative photomicrographs and quantification of positively stained cells as mean ± s.d. of n = 3 independent experiments are shown. j, 2'3'-cGAMP ELISA analysis of VIS IMR90 cells as in i .
Extended Data Fig. 2 Additional biological properties of VIS exerted by a variety of viruses.
a, SA-β-gal staining in human cell lines (RPE1, A549) infected with AAV, lentivirus, HCoV-NL63, and VSV as in Fig. 1g. b, SA-β-gal staining of human primary bronchial or nasal epithelial cells (HBEpC and HNEpC, respectively) infected with HCoV-NL63 and VSV. Mock infected cells as negative control. Quantification of positive cells for n = 3 independent experiments is shown as mean ± s.d.; scale bar = 100 µm. c, Quantification result of SA-β-gal-positive cells (RPE1, A549) after lentiviral infection at MOI as indicated are shown as mean percentage ± s.d. for n = 3 independent experiments. Note that MOI 50 was chosen for VIS induction. d, Quantification result of SA-β-gal-positive RPE1 cells after infection with VSV-ΔG*-CoV-S or VSV-ΔG*-CoV-2-S at MOI as indicated are shown as mean percentage ± s.d. for n = 3 independent experiments. Note that MOI 10 was chosen for VIS induction. e, SA-β-gal staining in human cell lines (RPE1, A549) and primary HNEpC infected with VSV-ΔG*-CoV-S, VSV-ΔG*-CoV-2-S, or VSV-ΔG*/empty vector (VSV-ΔG*-emp) as in Fig. 1g. NIH3T3 as ACE2-negative, infection-resistant control. f, Fluorescence detection of ROS in IMR90 after infection with retrovirus or VSV and treatment with NAC as indicated (upper panels). SA-β-gal staining and quantification of corresponding samples (lower panels). Mock infection or solvent treatment controls (UT) are shown. Representative photomicrographs and quantification are shown as mean percentages ± s.d. for n = 3 independent experiments. Scale bar for ROS and SA-β-gal = 100 µm; for γH2A.X (insets; pink dots reflect foci) 5 µm. g, Quantification of γH2A.X-positive IMR90 cells as in f. n = 3 independent experiments is shown as mean ± s.d. h, 2'3'-cGAMP ELISA analysis of IMR90 infected and treated with GS-441524 as in g. n = 8 independent experiments is shown as mean ± s.d. i, Viral RNA detection in the supernatant of primary nasal epithelial cells with exogenous hACE2 expression (HNEpC-hACE2) infected with SARS-CoV-2, at the indicated time-points. p53 shRNA (shp53) renders cells senescence–incapable, but, unlike treatment with 10 μM GS-441524 (GS), does not block viral replication. Data are shown as mean values + s.d. for n = 3 independent experiments. j, Relative viability of the indicated conditions, each compared to the corresponding untreated (UT) control of SARS-CoV-2-infected or mock-infected cells as in i, 72 h after infection. Data are shown as mean values + s.d. for n = 3 independent experiments. k, GSEA probing selected senescence-related gene sets66 by RNA-seq (GSE147507) analysis of NHBE, Calu-3, and A549 cells infected with SARS-CoV-2, compared to corresponding mock-infected controls. Positive NES indicates enrichment in virus-infected cells (dark grey bars), negative NES indicates downregulation in virus-infected cells (light grey bars). NES of FDR q < 0.05 are considered statistically significant and presented (for individual q-values, see Supplementary Information). Biological replicates comprise n = 7 control and n = 3 infected regarding NHBE, n = 3 for each condition regarding Calu-3, and n = 5 control and n = 3 infected regarding A549.
Extended Data Fig. 3 Vignettes of VIS in human COVID-19 lung specimens.
a, IHA of senescence markers in autopsy lung specimens from individuals without COVID-19 (non-COVID-19; with no obvious airway pathology) vs. patients with SARS-CoV-2-PCR-proven COVID-19. Representative photomicrographs and quantification of affected area as mean percentages of positive cells ± s.d. or mean percentage of positive area ± s.d (only for lipofuscin) of patients with COVID-19 (lipofuscin, p21CIP1, H3K9me3, BCL2L2, n = 5; IL-8, n = 9; SARS-CoV-2 RNA, n = 4) and individuals without COVID-19 (n = 2). Note that in situ hybridization of SARS-CoV-2 RNA was occasionally, but not consistently positive in COVID-19 samples at autopsy (2/4). Scale bars represent 50 µm. b, scRNA-sequencing-based expression analysis of CDKN2A and targets of navitoclax (BCL2L1 and BCL2L2) in bronchiolar protected specimen brushes (PS), and bronchoalveolar lavages (BL) of patients with COVID-19 with critical course of the disease (n = 4). UMAPs of cell types (top). Orange color code marks cells expressing CDKN2A, purple BCL2L1 (lower left) or BCL2L2 (lower right), and red for both. c, Gene expression profile (GEP) indicating elevated transcript expression of cytokines and markers related to M1 macrophage polarization and activation32,33 by oligonucleotide-based transcript hybridization of lung specimens as in a (n = 3 each). d, CD62/P-selectin-marked platelet activation by flow cytometry in whole blood samples of healthy donors (n = 7) or COVID-19 patients (n = 10). Mean ± s.d. with individual values as dots. *p = 0.0368 by unpaired t-test (two-tailed). e, Volcano plot of proteins differentially (p < 0.05) expressed in COVID-19 patient serum; SASP factor SERPINA367 highlighted in red as presented in Fig. 2g. Combined clinical COVID-19 severity WHO grade 3 and grade 4 (n = 86 patients) vs. grade 7 (n = 52 patients) are compared. The log2-transformed fold changes are plotted against log10 adjusted (Benjamini-Hochberg) p-values. Vertical red lines indicate log-transformed fold change cutoffs −0.5/0.5, horizontal red line indicates a significance cutoff p = 0.05. Analysis was performed using the DEqMS software package. f, Scatter plot showing average level of 16 SASP factors measured as in Fig. 2e and fibrinogen concentration in the serum (y-axis) of the same individual patients. n = 3 severe, n = 5 moderate, and n = 18 mild COVID-19 patients and healthy control (n = 6) are shown as color-coded dots. Correlation coefficiency (Pearson) r = 0.4139 with p = 0.0103. g, Expression levels of senescence-related transcripts (gene sets: Reactome_SASP and SASP_Schleich68) by oligonucleotide-based transcript hybridization of nasopharyngeal and lung specimens of patients with COVID-19 (n = 3) separated by their histopathological microthrombosis status. Shown are genes with fold-change > 1.5. h, Percentages of IL-8-positive cells in COVID-19 lung specimens as in a, now stratified by their microthrombosis status (positive, n = 4; negative n = 5), are shown as mean values + s.d.. p = 0.0397 by unpaired t-test (two-tailed).
Extended Data Fig. 4 Additional COVID-19-relevant biological features evoked by the VIS-associated secretome.
a, Percentage of CD13+ THP-1 macrophages exposed to SN from retroviral VIS vs. proliferating HDF as indicated and shown in Fig. 3a. n = 3 (n = 4 for medium control) independent experiments; shown are mean percentages + s.d. *p = 0.0017 for IMR90 and 0.0029 for WI38 by unpaired t-test (two-tailed). b, GSEA of SNVIS-primed macrophages unveiled vignettes of senescence and inflammation in the CD86+ population by probing of selected senescence- or macrophage activation-related GO terms and gene sets66,68 in RNA-seq analyses of THP-1 macrophages incubated with IMR90 SNVIS and sorted by CD86 expression status (as in Fig. 3c, left). n = 2 for each condition. Gene sets with FDR q ≤ 0.05 are shown. c, SA-β-gal staining (left) and gene expression analysis by RT-qPCR (right) of HUVEC endothelial cells that entered secondary, paracrine senescence upon exposure to SNVIS of WI38 as indicated. Representative photomicrographs and quantification of n = 3 independent experiments; shown are mean percentages ± s.d. Relative expression levels of core senescence-related transcripts (left panel) and SASP transcripts (right panel) are shown as mean values + s.d.
Extended Data Fig. 5 Targets of senolytic drugs are induced after viral infection and render VIS cells sensitive to senolytics.
a, Gene expression analysis of BCL2 family transcripts by RT-qPCR in VIS HDF, with mock infection as a control, as indicated. n = 3 independent experiments for each cell line and infection. b, STITCH plot showing interactions between senolytic drugs (navitoclax, fisetin, dasatinib, quercetin) and their potential targets. Protein-protein interactions are shown in grey, chemical-protein interactions in green and interactions between chemicals in red. Modified from http://stitch.embl.de. Note that all senolytics shown are working in close proximity and have overlapping targets at the level of kinase networks and BCL2-family members. c, Relative viability of proliferating or VIS (retrovirus) WI38 and Tig3 HDFs as in Extended Data Fig. 1a, 24 h after treatment with 1 µM venetoclax, 1 µM navitoclax, or solvent-only (DMSO). n = 3 independent experiments. d, Relative viability of senescence-incapable IMR90 cells (as in Fig. 1e, 4b) infected with VSV, and 24 h after treatment with 1 µM venetoclax, 1 µM navitoclax, 20 µM fisetin, 20 µM D/Q or solvent-only (DMSO). n = 3 independent experiments. e, Relative viability of proliferating or VIS IMR90 HDF compared to solvent-only (DMSO) control, two days after treatments with 1 µM navitoclax, 1 µM MCL1 inhibitor AT101, or both. Results shown as mean relative viability + s.d. for n = 3 independent experiments. Of note, strongly MCL-1-expressing IMR90 VIS cells (see Fig. 4a) were not more susceptible to combined treatment with navitoclax plus an MCL-1 inhibitor as compared to navitoclax alone. f, Secondary viral infectivity by SN transfer. Wild-type MEF that entered VIS in response to replication-competent VSV/GFP (or mock) were treated with 1 µM navitoclax before transferring the SN to uninfected THP-1 macrophages, here used as an infectivity reporter line. SA-β-gal staining to illustrate VIS, and GFP detection to visualize VSV infection of MEF. Secondary infectivity was measured as the GFP-positive fraction of THP-1 cells exposed to SN from MEF after navitoclax or solvent-only treatment (bar plot). Senescence-dependent killing of VIS MEF did not increase GFP-positive THP-1 cells, thereby demonstrating that non-assembled cytoplasmic virus material potentially released upon senolysis does not add to infectivity. n = 3 independent experiments. g, Secondary viral infectivity (% GFP) and relative viability (Rel. viability) of THP-1 cells after SN transfer from VSV- or mock-infected MEFs. Dilution of VIS SN as indicated below decreases secondary infection and increases viability. n = 3 independent experiments. h, Viability of HUVEC endothelial cells after exposure to human serum and SN of proliferating (SNprol.) or VIS (SNVIS) primary nasal epithelial cells (HNEpC-ACE2) as in Fig. 4f. n = 3 independent experiments. i – l, SARS-CoV-2 spike protein-pseudotyped VSV-ΔG*/CoV-2-S system to probe VIS in response to spike protein-affecting mutations. i, SA-β-gal staining of human cell line RPE1 infected with VSV-ΔG*-CoV-2-S (WT), VSV-ΔG*-CoV-2-S variant B.1.1.7 (Alpha), VSV-ΔG*-CoV-2-S variant B.1.351 (Beta), or mock infection as control. No obvious difference in the ability to drive VIS was detected. Representative photomicrographs and quantifications are shown as mean percentages ± s.d. for n = 3 independent experiments. Scale bar represents 100 µm. j, CD86-marked activation of THP-1 cells exposed to SN of RPE1 cells in VIS after infection with VSV-ΔG* pseudotypes as indicated, or mock infection, as in i. Fractions of CD86+ cells relative to mock are shown. SN of cells infected with either mutant- or wild-type-pseudotyped virus equally enhance the fraction of CD86+ THP-1 cells. n = 3 independent experiments. k, Viability of cells as in i, after senolytic treatments as indicated. n = 3 independent experiments. All senolytic regimens, navitoclax, fisetin and D/Q proved equally effective in selectively eliminating VIS cells induced by the wild-type or the alpha or beta mutants. l, Gene expression analysis of senescence-related genes by RT-qPCR in RPE1 cells as in i. While transcript profiles of cell-autonomous senescence markers and pro-coagulatory SASP factors were similar between wild-type and mutants, pro-inflammatory cytokines and extracellular matrix-active factors appeared to be significantly higher expressed in the two mutant scenarios compared to wild-type. n = 3 independent experiments. All bar plots in this figure show mean results + s.d. with individual values as dots. *p < 0.05 by unpaired t-test (two-tailed) in all panels of this figure.
Extended Data Fig. 6 Senolysis as a novel treatment concept in the COVID-19 golden hamster model.
a, scRNA-seq-based expression analysis of viral RNA and senescence-associated genes in SARS-CoV-2-infected Syrian golden hamster lungs (n = 3 per time-point). UMAPs of cell types (top left, cell types of special interest in red), core senescence genes (top right), Cdkn2a (bottom left), SASP genes (bottom second from left), and Src (bottom right) on the indicated day after infection.; orange color code marks cells expressing senescence-associated genes, purple viral RNA, and red both. b, scRNA-seq-based expression analysis of Bcl-w, Bcl2 and Mcl1 transcripts in the indicated cell types in SARS-CoV-2-infected Syrian golden hamster lungs (n = 3 per time-point) at the indicated days after infection.; average-scaled (z-scores) expression levels are shown in color code. Note that Bcl-2 and M-1 transcript levels followed no clear trend, while Bcl-w transcripts were induced at day 5 after infection in all cell types presented. c, Daily body weight assessment of Syrian golden hamsters infected with SARS-CoV-2 (or uninfected healthy controls), and subsequently treated with navitoclax or solvent-only as in Fig. 5a. n = 5 each for SARS-CoV-2-infected solvent-only- and navitoclax-treated animals; n = 3 for the healthy control group. d, Number of platelets per 500 erythrocytes in blood smears of golden hamsters infected with SARS-CoV-2 and treated as in c, on day 7 after infection. Mean counts ± s.d. of n = 5 hamsters per treatment group. *p = 0.0127 by unpaired t-test (two-tailed). Note for c and d: Although the navitoclax group presented with relative weight loss of around nine percent and a reduction of nearly 40% of the initial thrombocyte concentration compared to the infection-only group at the end of therapy, a prominent adverse effect of navitoclax, no obvious external or internal signs of enhanced hemorrhage was observed. e, Transmission electron microscopy analysis of trachea epithelial cells from hamsters as in b and c. In infected and solvent treated animals (with the enlarged photomicrograph of a nucleus on the right), ciliated epithelial cells show senescent features as dilated endoplasmic reticulum (*), disrupted and dilated nuclear envelope (N = nucleus), vacuolization and altered mitochondria. Scale bar = 1µm. n = 4 for SARS-CoV-2 infected groups and n = 3 for the mock infection group. f, Representative photomicrographs of H/E-stained lung tissue of Syrian golden hamsters as in Fig. 5a and c–e at day 7 after infection.. Lung sections for overview (i, v, ix) with active inflammation (arrow), signs of necrosis (inset, arrowhead), and hyperplasia (arrowheads) of alveolar epithelial cells (AEC) II (ii, vi). Vascular endothelialitis (iii, vii) with sloughing of endothelial cells (arrow), and subendothelial infiltration by lymphocytes (arrowhead). Regeneration of bronchial epithelium (iv, viii; double-headed arrow). Lung of mock-infected, untreated animals as control (ix, x, xi, xii). General signs of inflammation and AEC II hyperplasia in the navitoclax group were rather similar to untreated animals. while the navitoclax group presented with a less extensive patchy pneumonia pattern when compared to the solvent-only group (compare f-v to f–i). n = 5 each for SARS-CoV-2-infected solvent-only- and navitoclax-treated animals. n = 3 for the healthy control group. Scale bars in i, v, ix = 1 mm, ii-iv, vi-viii, x-xii = 50 µm, insets ii, vi, x = 20 µm. g, Quantification of the findings shown in f. *p = 0.0042 by unpaired t-test (two-tailed). h, Multiplex bead-based protein analysis of golden hamster serum samples at day 7 after infection.; hamsters as in c–g. All bar plots in this figure show mean results + s.d. with individual values as dots.
Extended Data Fig. 7 Senolysis as a novel treatment concept in COVID-19 in additional animal models.
a, Daily body weight assessment of dwarf hamsters and K18-hACE2 mice infected with SARS-CoV-2 (or uninfected healthy controls), and subsequently treated with senolytics as in b–e e and Fig. 5b–e. Dwarf hamsters: n = 5 each for SARS-CoV-2-infected solvent-only-, navitoclax- or D/Q-treated; n = 2 for the healthy control group. K18-hACE2-transgenic mice: n = 5 for solvent, navitoclax, and D/Q; n = 4 for fisetin. b, Survival rate of K18-hACE2-transgenic mice on day 6d after infection with SARS-CoV-2 and subsequent senolytic treatment as in a. c, Histological scoring of H/E-stained lung tissue of K18-hACE2 mice as in a and b. d, Viral load in hamster and mouse tissues on day 6 (mouse, as in a–c), day 7 (golden hamster, as in Fig. 5a and Extended Data Fig 6), and day 4 (dwarf hamster, as in Fig. 5b-e) after infection. e, IHA of senescence markers (p16INK4A, H3K9me3) and lipofuscin-staining in lung specimens of K18-hACE2-transgenic mice as in a–d. All bar plots in this figure show mean results + s.d. with individual values as dots.
Extended Data Fig. 8 Senolysis as a novel treatment concept in COVID-19 patients.
a, Frequency (bars) and risk (odds ratio) of COVID-19 patients to be hospitalized or in need of oxygen support after either standard care (SC) or quercetin plus standard care (Quercetin). n = 97 for each group, based on two randomized trials combined and re-analysed here51,52. b, Days of hospitalization (left) of COVID-19 patients (left; *p < 0.0001 by unpaired t-test, two-tailed), the fraction of patients admitted to the intensive care unit (ICU; middle) or deceased in the course of the disease (right). Note that there was no patient in ICU or deceased in the quercetin group. Patients as in a; n = 97 for each group.
Supplementary information
Supplementary Information
This file contains details of the experimental procedures, Supplementary Tables 1 and 2, Supplementary References and Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Lee, S., Yu, Y., Trimpert, J. et al. Virus-induced senescence is a driver and therapeutic target in COVID-19. Nature 599, 283–289 (2021). https://doi.org/10.1038/s41586-021-03995-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03995-1
This article is cited by
-
Activation of senescence in critically ill patients: mechanisms, consequences and therapeutic opportunities
Annals of Intensive Care (2024)
-
Human cytomegalovirus infection triggers a paracrine senescence loop in renal epithelial cells
Communications Biology (2024)
-
COVID-19 drug discovery and treatment options
Nature Reviews Microbiology (2024)
-
DNA damage response(DDR): a link between cellular senescence and human cytomegalovirus
Virology Journal (2023)
-
Impaired autophagy-accelerated senescence of alveolar type II epithelial cells drives pulmonary fibrosis induced by single-walled carbon nanotubes
Journal of Nanobiotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.