Abstract
The enzymes of the mitochondrial electron transport chain are key players of cell metabolism. Despite being active when isolated, in vivo they associate into supercomplexes1, whose precise role is debated. Supercomplexes CIII2CIV1-2 (refs. 2,3), CICIII2 (ref. 4) and CICIII2CIV (respirasome)5,6,7,8,9,10 exist in mammals, but in contrast to CICIII2 and the respirasome, to date the only known eukaryotic structures of CIII2CIV1-2 come from Saccharomyces cerevisiae11,12 and plants13, which have different organization. Here we present the first, to our knowledge, structures of mammalian (mouse and ovine) CIII2CIV and its assembly intermediates, in different conformations. We describe the assembly of CIII2CIV from the CIII2 precursor to the final CIII2CIV conformation, driven by the insertion of the N terminus of the assembly factor SCAF1 (ref. 14) deep into CIII2, while its C terminus is integrated into CIV. Our structures (which include CICIII2 and the respirasome) also confirm that SCAF1 is exclusively required for the assembly of CIII2CIV and has no role in the assembly of the respirasome. We show that CIII2 is asymmetric due to the presence of only one copy of subunit 9, which straddles both monomers and prevents the attachment of a second copy of SCAF1 to CIII2, explaining the presence of one copy of CIV in CIII2CIV in mammals. Finally, we show that CIII2 and CIV gain catalytic advantage when assembled into the supercomplex and propose a role for CIII2CIV in fine tuning the efficiency of electron transfer in the electron transport chain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Structures of the CIII2CIV intermediate, assembled locked, mature unlocked and CIII2 from CICIII2 were deposited in the PDB (access IDs 7O3E, 7O37, 7O3C and 7O3H, respectively) and corresponding cryo-electron microscopy density maps in the EMDB (IDs 12705, 12702, 12703 and 12706, respectively). Any other relevant data and information are available from the corresponding author on request. The following models, deposited in the PDB (https://www.rcsb.org), have been cited throughout the paper: 1NTZ, 3CX5, 3L75, 5IY5, 5J4Z, 5Z62, 6HWH, 6HU9, 6QBX and 7JRP.
References
Schägger, H. & Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000).
Calvo, E. et al. Functional role of respiratory supercomplexes in mice: SCAF1 relevance and segmentation of the Qpool. Sci. Adv. 6, eaba7509 (2020).
Lobo-Jarne, T. et al. Human COX7A2L regulates complex III biogenesis and promotes supercomplex organization remodeling without affecting mitochondrial bioenergetics. Cell Rep. 25, 1786-1799.e4 (2018).
Letts, J. A., Fiedorczuk, K., Degliesposti, G., Skehel, M. & Sazanov, L. A. Structures of respiratory supercomplex I+III2 reveal functional and conformational crosstalk. Mol. Cell 75, 1131-1146.e6 (2019).
Lobo-Jarne, T. & Ugalde, C. Respiratory chain supercomplexes: structures, function and biogenesis. Semin. Cell Dev. Biol. 76, 179–190 (2018).
Davies, K. M., Blum, T. B. & Kühlbrandt, W. Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl Acad. Sci. USA 115, 3024–3029 (2018).
Letts, J. A., Fiedorczuk, K. & Sazanov, L. A. The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016).
Sousa, J. S., Mills, D. J., Vonck, J. & Kühlbrandt, W. Functional asymmetry and electron flow in the bovine respirasome. eLife 5, e21290 (2016).
Gu, J. et al. The architecture of the mammalian respirasome. Nature 537, 639–643 (2016).
Guo, R., Zong, S., Wu, M., Gu, J. & Yang, M. Architecture of human mitochondrial respiratory megacomplex I 2 III 2 IV 2. Cell 170, 1247–1257 (2017).
Hartley, A. M. et al. Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc 1. Nat. Struct. Mol. Biol. 26, 78–83 (2019).
Rathore, S. et al. Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 26, 50–57 (2019).
Maldonado, M., Guo, F. & Letts, J. A. Atomic structures of respiratory complex III2, complex IV, and supercomplex III2-IV from vascular plants. eLife 10, e62047 (2021).
Cogliati, S. et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582 (2016).
Spikes, T. E., Montgomery, M. G. & Walker, J. E. Structure of the dimeric ATP synthase from bovine mitochondria. Proc. Natl Acad. Sci. USA 117, 23519–23526 (2020).
Pinke, G., Zhou, L. & Sazanov, L. A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 27, 1077–1085 (2020).
Wiseman, B. et al. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat. Struct. Mol. Biol. 25, 1128–1136 (2018).
García‐Poyatos, C. et al. Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep. 21, e50287 (2020).
Lopez-Fabuel, I. et al. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl Acad. Sci. USA 113, 13063–13068 (2016).
Xia, D. et al. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim. Biophys. Acta 1827, 1278–1294 (2013).
Pérez-Pérez, R. et al. COX7A2L is a mitochondrial complex III binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep. 16, 2387–2398 (2016).
Zong, S. et al. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018).
Zong, S. et al. UQCRFS1N assembles mitochondrial respiratory complex-III into an asymmetric 21-subunit dimer. Protein Cell 9, 586–591 (2018).
Hunte, C., Koepke, J., Lange, C., Roßmanith, T. & Michel, H. Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure 8, 669–684 (2000).
Stuchebrukhov, A., Schäfer, J., Berg, J. & Brzezinski, P. Kinetic advantage of forming respiratory supercomplexes. Biochim. Biophys. Acta Bioenerg. 1861, 148193 (2020).
Berndtsson, J. et al. Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. EMBO Rep. 21, e51015 (2020).
Blanchi, C., Genova, M. L., Castelli, G. P. & Lenaz, G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J. Biol. Chem. 279, 36562–36569 (2004).
Lapuente-Brun, E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013).
Moe, A., Di Trani, J., Rubinstein, J. L. & Brzezinski, P. Cryo-EM structure and kinetics reveal electron transfer by 2D diffusion of cytochrome c in the yeast III–IV respiratory supercomplex. Proc. Natl Acad. Sci. USA 118, e2021157118 (2021).
Fedor, J. G. & Hirst, J. Mitochondrial supercomplexes do not enhance catalysis by quinone channeling. Cell Metab. 28, 525-531.e4 (2018).
Letts, J. A., Degliesposti, G., Fiedorczuk, K., Skehel, M. & Sazanov, L. A. Purification of ovine respiratory complex i results in a highly active and stable preparation. J. Biol. Chem. 291, 24657–24675 (2016).
Kampjut, D., Steiner, J. & Sazanov, L. A. Cryo-EM grid optimisation for membrane proteins. iScience 24, 102139 (2021).
Cheng, A. et al. High resolution single particle cryo-electron microscopy using beam-image shift. J. Struct. Biol. 204, 270–275 (2018).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Sherwood, S. & Hirst, J. Investigation of the mechanism of proton translocation by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria: does the enzyme operate by a Q-cycle mechanism? Biochem. J. 400, 541–550 (2006).
Kubota, T., Yoshikawa, S. & Matsubara, H. Kinetic mechanism of beef heart ubiquinol: cytochrome c oxidoreductasel. J. Biochem. 111, 91–98 (1992).
Rieske, J. S. [44] Preparation and properties of reduced coenzyme Q-cytochrome c reductase (complex III of the respiratory chain). Methods Enzymol. 10, 239–245 (1967).
Trumpower, B. L. & Edwards, C. A. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate•cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem. 254, 8697–8706 (1979).
[No authors listed] Reduced cytochrome c solution. Cold Spring Harb. Protoc. 2015, pdb.rec084475 (2015).
Acknowledgements
We thank the pre-clinical facility of the IST Austria and A. Venturino for assistance with the animals; and V.-V. Hodirnau for assistance during the Titan Krios data collection, performed at the IST Austria. The data processing was performed at the IST high-performance computing cluster. This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 754411.
Author information
Authors and Affiliations
Contributions
I.V. purified the sample, prepared cryo-electron microscopy grids, acquired and processed EM data, built and analysed the atomic models and wrote the initial draft of the manuscript. L.A.S. designed and supervised the project, analysed data and models and revised the manuscript. I.V. and L.A.S. acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Werner Kühlbrandt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sample preparation overview.
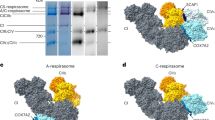
a, Purification of the mouse sample: on top, chromatograms of the MonoQ (left), the first SEC (middle) and the second SEC (right). The pooled fractions are highlighted in blue. At the bottom, native PAGE and western blot of the purification steps: from left to right, Coomassie-stained solubilised material (SM), then Coomassie staining, complex I in-gel activity assay, complex IV in-gel activity assay, western blot for Core2, western blot for SCAF1 of the purification steps. b, Purification of the ovine sample: the same steps as shown in a. For Gel source data, please see Supplementary Fig. 1. The mouse purification was repeated at least 5 times, the ovine preparation was repeated at least 10 times, all attempts of replication were successful, and one representative preparation per sample type is shown in the figure.
Extended Data Fig. 2 Processing pipeline for the mouse CIII2CIV dataset.
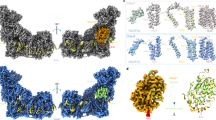
Scheme of the processing pipeline, with representative micrograph, pre-processing, classification steps and final maps, coloured by local resolution, with relative angular distribution plot.
Extended Data Fig. 3 Processing pipeline for the mouse CICIII2 data, FSC curves of the mouse maps and representative densities from the cryo-EM maps.
a, Processing pipeline as in Extended Data Fig. 2: the same set of micrographs was used, but the classification focused on CICIII2. b, Gold-standard half-map and model-map FSC curves for the mouse dataset. c–h, Representative densities of CIII2 (c–e) and CIV (f–i) for α-helices -contoured at 2.7σ- (c and f), β-sheets -contoured at 2.7σ- (d and g), cofactors -contoured at 1.9σ- (e and i) and lipids -contoured at 1.8σ- (h, from CIII2).
Extended Data Fig. 4 Processing pipeline and maps of the ovine CIII2CIV dataset.
a, Processing pipeline of the ovine CIII2CIV dataset, as in Extended Data Figs. 2, 3. b, Final maps of the ovine CIII2CIV locked (left, 2.8σ contour) and unlocked (right, 3σ contour for CIII and 6σ for CIV), shown from the side (top) and from the matrix (bottom). The respective mouse models, coloured as in Figs. 1, 3 , were superimposed on CIII2, to show the conservation of the conformations across mammalian species.
Extended Data Fig. 6 MPP cavity in different supercomplexes.
a, Cryo-EM map density (grey mesh) carved around Sub9 (orange cartoon) and SCAF1 (blue) positions in supercomplexes CIII2CIV, CICIII2 and CICIII2CIV. For CIII2CIV and the mouse CICIII2, the unique positions of a single copy each of Sub9 and SCAF1 were used, while for respirasomes, where the limited resolution prevented Sub9-focused classification and so CIII2 adopts a random orientation, two-fold symmetry-related positions of these proteins were also added. Contour levels are 2, 1.9 and 2.1σ for mouse CIII2CIV mature unlocked, assembled locked and intermediate locked respectively; 8σ for ovine CIII2CIV mature unlocked; 2.9σ for mouse CICIII2; 6.7σ for mouse CICIII2CIV; 6.7 and 5.3σ for ovine CICIII2CIV tight and loose respectively. b, Sub9 (orange) integration into the MPP walls: UQCRC1 (Core1) and UQCRC2 (Core2) are shown in grey (Core1 light and Core2 dark), except for the strands of Core2 forming continuous sheets with Sub9, which are shown in yellow. c, SCAF1 (blue), Sub7 (yellow) and ISP (red) arrangement in the intermediate (left) and assembled locked (right) classes. The rest of CIII is depicted in grey. The density for Sub7 is carved in grey, to show the ordering of the loop in the assembled versus intermediate class. In the insets at the bottom, the densities for SCAF1 and ISP are shown in the two classes. 2.9σ contour for CIII2CIV intermediate and 2.3σ for assembled.
Extended Data Fig. 7 SCAF1 in CIII and CIV and functional implications.
a, Surface charge representation (as in Fig. 3) of Core1 and SCAF1 in the unlocked class (full model is grey with a sketched eye indicating the perspective). SCAF1 cartoon is green. A white asterisk indicates the MPP "mouth". b, SCAF1 (cyan) and COX7A1 (blue) models fitted to the locked (left, 2.5σ contour) and unlocked class (right, 2.7σ contour) CIV maps. The arrows in b and c indicate bulky residues used to identify the subunit based on the map density. SCAF1 model, but not COX7A1, fits the density in both classes. c, Ovine SCAF1 (cyan) and COX7A1 (blue) models fitted to CIV map from respirasome (7.3σ contour): contrary to CIII2CIV, COX7A1 model fits the density, unlike SCAF1. d, Sequence alignment of mammalian COX7A isoforms. The SCAF1 portions interacting with CIII2 are indicated above the alignment. The black arrows indicate the positions highlighted in b and c. e, Activity measurements (described in the Methods) of CIII2+CIV (light grey) and supercomplex CIII2CIV (dark grey). The electron transfer rates (in e-/s) are calculated from the CIII2-driven oxidation of DQH2 or from the CIV-driven oxidation of reduced cyt c, as indicated. The bars represent the mean, the dots depict the single measurements. The 5-10 nM ox cyt c represents 2.5-5:1 cyt c/CIII2CIV ratio (two independent experiments). All other measurements come from 3 independent experiments (two values in the CIII2+CIV sample with 50 μM red cyt c overlap, therefore only two dots are visible). f, Cofactor distances within and between CIII2 and CIV, in the unlocked conformation. Cyt c is bound to CIII2 and CIV as in Fig. 3b. The transparent cartoon represents the protein subunits, the cofactors are sticks (hemes) and spheres (FeS clusters and Cu centers), in orange for CIII2, cyan for CIV and black for cyt c. The dashed lines represent the IMM.
Extended Data Fig. 8 Structural differences among supercomplexes and comparison between CIII2 movements in CICIII2 and intermediate vs assembled CIII2CIV.
a, Top left, CIII2CIV and respirasome (ovine, PDB 5j4z) aligned by CIII2: CI of respirasome is grey, CIV of respirasome is blue, CIII2 is yellow and CIV of CIII2CIV is cyan (the colour scheme for mammalian CIII2CIV is consistent throughout the panel). CIII2CIV locked is shown on the left, unlocked on the right. The two possible orientations of CIII2CIV based on CIII2 are shown at the top and at the bottom. Top right, plant CIII2CIV (PDB 7jrp) and mammalian CIII2CIV aligned by CIII2 in the locked (top) and unlocked (bottom) conformations. The plant CIV is blue. Bottom left, yeast CIII2CIV2 (PDB 6hu9) and mammalian CIII2CIV aligned by CIII2 in the locked (left) and unlocked (right) conformations. The yeast CIV is blue. Bottom right, M. smegmatis CIII2CIV2 (PDB 6hwh) and mammalian CIII2CIV aligned by CIII2 in the locked (left) and unlocked (right) conformations. The bacterial CIV is blue. The straight arrows indicate the shift of CIV towards mammalian CIV, and the curved arrows indicate the 90 or 180o flip of CIV around its vertical axis. b, Side (top) and IMS (bottom) views of the extreme frames of CIII2 3DVA mode 0 (open on the left and closed in the middle, 5σ contour). Ovals indicate the position of the ISP Fe-S domains, only visible in the closed state. On the right, overlay of the two extremes, with closed in dark grey and open in light grey. Arrows indicate the “breathing” movement of the TM and IMS domains. c, Same as b, but with CIII2 intermediate (left) and assembled (right) models from CIII2CIV fitted to the densities and coloured as in Figs. 1, 3.
Supplementary information
Supplementary Information
This file contains additional discussion relevant for the main text.
41586_2021_3927_MOESM4_ESM.jpg
Supplementary Fig. 1 Uncropped gels. All the original, uncropped scans of the native gels and western blots shown in Extended Data Fig. 1 are shown.
41586_2021_3927_MOESM5_ESM.mov
Supplementary Video 1 Overview of the structural features of CIII2CIV.The mature unlocked state map and model are shown, followed by a transition through the three different conformations, viewed from the side and from the top. Finally, the cytochrome c binding sites, inferred from the crystal structures cited in the manuscript, are shown. The colour scheme is the same as in the rest of the manuscript.
41586_2021_3927_MOESM6_ESM.mov
Supplementary Video 2 “Breathing” movement of CIII2 in CICIII2. The result of the 3DVA analysis on CIII2 is shown, viewed from the side first, then from the IMS. The fluctuating movements between a closed state, with the two ISP C-terminal domains visible, and an open state, with no density corresponding to ISP, are depicted.
Rights and permissions
About this article
Cite this article
Vercellino, I., Sazanov, L.A. Structure and assembly of the mammalian mitochondrial supercomplex CIII2CIV. Nature 598, 364–367 (2021). https://doi.org/10.1038/s41586-021-03927-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03927-z
This article is cited by
-
SCAF1 drives the compositional diversity of mammalian respirasomes
Nature Structural & Molecular Biology (2024)
-
Euglena’s atypical respiratory chain adapts to the discoidal cristae and flexible metabolism
Nature Communications (2024)
-
Electron transport system supercomplexes affect reactive-oxygen species production and respiration in both a hibernator (Ictidomys tridecemlineatus) and a nonhibernator (Rattus norvegicus)
Journal of Comparative Physiology B (2024)
-
A FRET-based respirasome assembly screen identifies spleen tyrosine kinase as a target to improve muscle mitochondrial respiration and exercise performance in mice
Nature Communications (2023)
-
Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.