Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of cancer deaths worldwide1. Studies in human tissues and in mouse models have suggested that for many cancers, stem cells sustain early mutations driving tumour development2,3. For the pancreas, however, mechanisms underlying cellular renewal and initiation of PDAC remain unresolved. Here, using lineage tracing from the endogenous telomerase reverse transcriptase (Tert) locus, we identify a rare TERT-positive subpopulation of pancreatic acinar cells dispersed throughout the exocrine compartment. During homeostasis, these TERThigh acinar cells renew the pancreas by forming expanding clones of acinar cells, whereas randomly marked acinar cells do not form these clones. Specific expression of mutant Kras in TERThigh acinar cells accelerates acinar clone formation and causes transdifferentiation to ductal pre-invasive pancreatic intraepithelial neoplasms by upregulating Ras–MAPK signalling and activating the downstream kinase ERK (phospho-ERK). In resected human pancreatic neoplasms, we find that foci of phospho-ERK-positive acinar cells are common and frequently contain activating KRAS mutations, suggesting that these acinar regions represent an early cancer precursor lesion. These data support a model in which rare TERThigh acinar cells may sustain KRAS mutations, driving acinar cell expansion and creating a field of aberrant cells initiating pancreatic tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data for KRAS variants is available with the SRA accession: PRJNA598774. All other data are available from the authors upon reasonable request. Source data are provided with this paper.
Code availability
Custom Python scripts used in this manuscript are available at (https://github.com/cmroake/KRAS_scripts).
References
Rawla, P., Sunkara, T. & Gaduputi, V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J. Oncol. 10, 10–27 (2019).
Jan, M. et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4, 149ra118 (2012).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Murtaugh, L. C. & Keefe, M. D. Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 77, 229–249 (2015).
Desai, B. M. et al. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet β cell, regeneration. J. Clin. Invest. 117, 971–977 (2007).
Westphalen, C. B. et al. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell 18, 441–455 (2016).
Sangiorgi, E. & Capecchi, M. R. Bmi1 lineage tracing identifies a self-renewing pancreatic acinar cell subpopulation capable of maintaining pancreatic organ homeostasis. Proc. Natl Acad. Sci. USA 106, 7101–7106 (2009).
Hruban, R. H., Goggins, M., Parsons, J. & Kern, S. E. Progression model for pancreatic cancer. Clin. Cancer Res. 6, 2969–2972 (2000).
Kopp, J. L. et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012).
Habbe, N. et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl Acad. Sci. USA 105, 18913–18918 (2008).
De La, O. J. et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc. Natl Acad. Sci. USA 105, 18907–18912 (2008).
Flowers, B. M. et al. Cell of origin influences pancreatic cancer subtype. Cancer Discov. 11, 660–677 (2021).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Lee, J. et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat. Commun. 8, 14686 (2017).
Kanda, M. et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142, 730–733.e739 (2012).
Shi, C. et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol. Cancer Res. 7, 230–236 (2009).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Ying, H. et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 30, 355–385 (2016).
Roake, C. M. & Artandi, S. E. Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 21, 384–397 (2020).
Pech, M. F. et al. High telomerase is a hallmark of undifferentiated spermatogonia and is required for maintenance of male germline stem cells. Genes Dev. 29, 2420–2434 (2015).
Petersen, G. M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Lin, S. et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018).
Morris, J. P. T., Cano, D. A., Sekine, S., Wang, S. C. & Hebrok, M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 120, 508–520 (2010).
Pylayeva-Gupta, Y., Grabocka, E. & Bar-Sagi, D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 (2011).
Collins, M. A., Yan, W., Sebolt-Leopold, J. S. & Pasca di Magliano, M. MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology 146, 822–834.e827 (2014).
Bryant, K. L., Mancias, J. D., Kimmelman, A. C. & Der, C. J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci 39, 91–100 (2014).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181 (2012).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Kopinke, D. et al. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev. Biol. 362, 57–64 (2012).
Hegyi, P. et al. l-arginine-induced experimental pancreatitis. World J. Gastroenterol. 10, 2003–2009 (2004).
Acknowledgements
This work was supported by grants from the NIH (AG056575, CA244114 and CA197563; S.E.A.), the Emerson Collective (S.E.A.), anonymous support (S.E.A.), the DFG (NE 2006/1-1; P.N.), California TRDRP (25FT-0011; P.N.) and the California Institute for Regenerative Medicine (R.J.L.). We thank members of the Artandi laboratory for critical comments and P. Chu from the Stanford Human Pathology/Histology Service Center for technical assistance, and acknowledge the Stanford Functional Genomics Facility and Stanford Shared FACS Facility.
Author information
Authors and Affiliations
Contributions
P.N. and S.E.A. conceived the study. P.N. and S.E.A. designed the experiments. P.N., S.J.K. and R.J.L. performed histological analysis. P.N. and R.B.W. performed the LCM experiments. G.W.C. analysed and evaluated the human patient samples. C.M.R. performed the sequencing analyses. P.N. and S.E.A. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Laura Wood and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
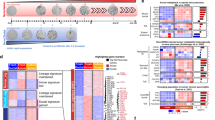
Extended Data Fig. 1 Identification of a TERT-high subpopulation in acinar cells and beta cells but not duct cells using lineage tracing.
a, TertCreERT2/+; Rosa26LSL-tdTomato/+ mouse, which responds to tamoxifen-induced Cre-mediated activation of the tdTomato reporter allele. b, Immunohistochemistry analysis of Tomato in a TertCreERT2/+; Rosa26LSL-tdTomato/+ (TertCreER) pancreas after oil and tamoxifen injection (n=4 mice for oil ctrl; n=11 mice for tamoxifen). Mo, month. c, d, Co-immunofluorescence stains 10 days after single dose of tamoxifen (5mg) in TertCreER pancreas (n=4 mice). Tomato (red, c, d), DAPI (blue, c, d), Insulin (grey, c, d) Sox9 (green, c) and Amylase (green, d) are shown. e, f, Single-molecule RNA FISH for Tert mRNA (e) on FACS-purified bulk acinar cells (e, f) from TertCreER mice 10d after Tamoxifen (5mg) injection (n=4 mice). g, Quantification of Tomato-positive cells in TertCreER mice in the acinar cell compartment 10 days and within pancreatic ductal cells 10 days and 12 months post tamoxifen activation (n=5 mice). h, qRT-PCR for indicated genes in FACS-sorted TomNeg (white bars) and TomPos (grey bars) acinar cells. i, j, Immunofluorescence analysis of a TertCreER pancreas 10 days after a single dose of 5mg tamoxifen (n=5 mice). Tomato (red), Dclk1 (i, green) or BMI1 (j, green) and DAPI (blue) are shown. Scale bar, 100µm (b), 50µm (e, f) and 25µm (c, d, i, j). All data are mean± SD. P values calculated by two-sided Mann-Whitney test (h).
Extended Data Fig. 2 Randomly marked acinar cells show limited clonogenicity and TERT-high acinar cells renew to yield TERT-low cells.
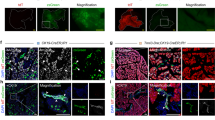
a, Schematic of lineage tracing in TertCreER mice treated with a single dose of tamoxifen on postnatal day 60 and analysis at indicated time points. Mo, month. b–d, Immunohistochemistry for Tomato in Ptf1aCreER mice 10d post tamoxifen (5mg (b), 0.5mg (c) and 0.05mg (d)) injection (n=4 mice). e, Distribution of Tomato-positive acinar cells per clone for each time point in Ptf1aCreER mice (each color represents one mouse; n=4 for each time point) f, TertCreER mice are injected with Tamoxifen (5mg) on postnatal day 60 and put on BrdU water (1mg/ml) for 30 days. g, Immunofluorescence analysis of a TertCreER pancreas after 30 days of BrdU water. Tomato (red), BrdU (green) and DAPI (blue) are shown. TERTHigh Tomato/BrdU double positive cells (green arrow) and TERTLow BrdU positive cells (orange arrow) shown. h, Quantification of TERTLow and TERTHigh BrdU+ cells (n=5 mice) (mean ± SD). i–k, Single-molecule RNA FISH for Tert on FACS-purified bulk (i), TERTHigh (j) and TERTLow derived (k) acinar cells of TertCreER mice(n=3). Scale bars, 25µm (g), 50µm (i–k), 100 µm (b–d). P values calculated by two-sided Mann-Whitney test (h).
Extended Data Fig. 3 During injury, TERTHigh acinar show enhanced repopulation compared with randomly marked acinar cells.
a, Schema of cerulein-induced pancreatitis. b, Immunofluorescence analysis of TertCreER mice 30 days after cerulein injections. Tomato (red), Ki67 (green) and DAPI (blue) are shown. TERTHigh, Ki67+ positive cells (green arrow) and TERTLow, Ki67+ cells (orange arrows) are shown. c, Quantification of TERTHigh, Ki67+ and TERTLow, Ki67+ cells at indicated time points (n=5 mice) (mean ± SD). d, Schematic of L-Arginine induced pancreatitis. e, Immunofluorescence analysis of TertCreER mice 30 days after L-Arginine induced pancreatitis. f, Quantification of Tomato-positive cells per clone with and without injury (n=5 mice) (mean ± SD). Scale bars, 25µm (d), 250 µm (e). P values calculated by two-sided Mann-Whitney test (c, f).
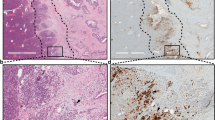
Extended Data Fig. 4 Enhanced PanIN formation from TERT-High acinar cells compared with sparsely labeled randomly marked acinar cells.
a, Schema for cerulein-induced pancreatitis. b, Immunohistochemistry for Tomato in Ptf1aCreER; Kras mice and TertCreER; Kras mice one month after cerulein-induced pancreatitis (AP). c, Quantification of number of clones per 20x high-power field (HPF) in Ptf1aCreER; Kras mice (n=7) and TertCreER; Kras mice (n=7) one month after cerulein-induced pancreatitis (AP) (mean ± SD). d, e, Representative metaplastic area (d, dashed line) and PanIN lesion (e, black arrow) in H&E staining of TertCreER; Kras mice one-month post injury (AP) (n=9 mice). f, g, Representative H&E staining (f) and Immunohistochemistry for Tomato (g) of Ptf1aCreER; Krasmice one-month post injury (AP) with high dose tamoxifen (5mg) (n=9 mice). h–k, Immunofluorescence analysis of a TertCreER (h, i) and TertCreER; Kras (j, k) mice one month after sham (h, j) or cerulein (i, k) treatment. Acinar cell expansion (red arrows) and PanINs (white arrow) are shown. Scale bar 100µm (a, c–f) and 50µm (g–j).
Extended Data Fig. 5 MEK inhibitor prevents PanIN formation from TERT-High cells expressing mutant Kras.
a, Schema of cerulein-induced pancreatitis with MEKi treatment (Trametinib). b, c, H&E (b) and Tomato (c) staining of TertCreER; Kras mice one month after cerulein induced pancreatitis with (Trametinib) and without (Vehicle) MEKi treatment. d, Quantification of Tomato-positive cells per clone with (n=4 mice) and without (n=3 mice) MEK inhibition (mean ± SD). Scale bars, 100µm.
Extended Data Fig. 6 Specificity of pERK-positive acinar regions in human samples.
pERK 1/2 staining of human pancreas cancer specimen using indicated antibodies and IgG control (n=7 for #4376, n=44 for #4370 and n=3 for IgG control. Scale bars, 100µm.
Supplementary information
Rights and permissions
About this article
Cite this article
Neuhöfer, P., Roake, C.M., Kim, S.J. et al. Acinar cell clonal expansion in pancreas homeostasis and carcinogenesis. Nature 597, 715–719 (2021). https://doi.org/10.1038/s41586-021-03916-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03916-2
This article is cited by
-
Reconstitution of human PDAC using primary cells reveals oncogenic transcriptomic features at tumor onset
Nature Communications (2024)
-
Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review
Signal Transduction and Targeted Therapy (2023)
-
Telomerase is required for glomerular renewal in kidneys of adult mice
npj Regenerative Medicine (2022)
-
Pancreatic cancer: why the cell of origin matters
Nature Reviews Gastroenterology & Hepatology (2022)
-
Association between cancer genes and germ layer specificity
Medical Oncology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.