Abstract
Atherosclerotic cardiovascular disease causes heart attacks and strokes, which are the leading causes of mortality worldwide1. The formation of atherosclerotic plaques is initiated when low-density lipoproteins bind to heparan-sulfate proteoglycans (HSPGs)2 and become trapped in the subendothelial space of large and medium size arteries, which leads to chronic inflammation and remodelling of the artery wall2. A proliferation-inducing ligand (APRIL) is a cytokine that binds to HSPGs3, but the physiology of this interaction is largely unknown. Here we show that genetic ablation or antibody-mediated depletion of APRIL aggravates atherosclerosis in mice. Mechanistically, we demonstrate that APRIL confers atheroprotection by binding to heparan sulfate chains of heparan-sulfate proteoglycan 2 (HSPG2), which limits the retention of low-density lipoproteins, accumulation of macrophages and formation of necrotic cores. Indeed, antibody-mediated depletion of APRIL in mice expressing heparan sulfate-deficient HSPG2 had no effect on the development of atherosclerosis. Treatment with a specific anti-APRIL antibody that promotes the binding of APRIL to HSPGs reduced experimental atherosclerosis. Furthermore, the serum levels of a form of human APRIL protein that binds to HSPGs, which we termed non-canonical APRIL (nc-APRIL), are associated independently of traditional risk factors with long-term cardiovascular mortality in patients with atherosclerosis. Our data reveal properties of APRIL that have broad pathophysiological implications for vascular homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA sequencing datasets (from vascular smooth muscle cells) are available in the Gene Expression Omnibus with accession codes GSE117963 and GSE17858. All other relevant data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
Code availability
For the clinical studies, the calculations were performed with SPSS (version 20.0, SPSS Inc.) for Windows, R version 3.6.0 (https://www.R-project.org/), and survival analysis was performed using the R packages survival (https://CRAN.R-project.org/package=survival) and survminer (https://CRAN.R-project.org/package=survminer). For analysis of mass spectrometry data, acquired raw data files were processed using Proteome Discoverer 2.4.1.15 SP1 for DDA experimental data or Skyline version 20.1.0.155 for PRM experimental data or using Mascot version 2.3.02 (Matrix Science, London, UK) and Phenyx (GeneBio, Geneva, Switzerland) as search engines. RNA-seq data were quality controlled using FastQC v0.11.3 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and trimmed using the Trim Galore v0.4.1 wrapper (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads were aligned to the GRCm38 mouse reference genome using Tophat v2.0.12. Reads with a minimum map quality of 20 were imported into Seqmonk 1.45.4 (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk).
References
Libby, P. The changing landscape of atherosclerosis. Nature 592, 524–533 (2021).
Gisterå, A. & Hansson, G. K. The immunology of atherosclerosis. Nat. Rev. Nephrol. 13, 368–380 (2017).
Ingold, K. et al. Identification of proteoglycans as the APRIL-specific binding partners. J. Exp. Med. 201, 1375–1383 (2005).
Vincent, F. B., Morand, E. F., Schneider, P. & Mackay, F. The BAFF/APRIL system in SLE pathogenesis. Nat. Rev. Rheumatol. 10, 365–373 (2014).
Castigli, E. et al. Impaired IgA class switching in APRIL-deficient mice. Proc. Natl Acad. Sci. USA 101, 3903–3908 (2004).
Huard, B. et al. APRIL secreted by neutrophils binds to heparan sulfate proteoglycans to create plasma cell niches in human mucosa. J. Clin. Invest. 118, 2887–2895 (2008).
McCarron, M. J., Park, P. W. & Fooksman, D. R. CD138 mediates selection of mature plasma cells by regulating their survival. Blood 129, 2749–2759 (2017).
Hymowitz, S. G. et al. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J. Biol. Chem. 280, 7218–7227 (2005).
Sandberg, W. J. et al. The tumour necrosis factor superfamily ligand APRIL (TNFSF13) is released upon platelet activation and expressed in atherosclerosis. Thromb. Haemost. 102, 704–710 (2009).
Lord, M. S. et al. The multifaceted roles of perlecan in fibrosis. Matrix Biol. 68-69, 150–166 (2018).
Tran-Lundmark, K. et al. Heparan sulfate in perlecan promotes mouse atherosclerosis: roles in lipid permeability, lipid retention, and smooth muscle cell proliferation. Circ. Res. 103, 43–52 (2008).
Sarrazin, S., Lamanna, W. C. & Esko, J. D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 (2011).
Parish, C. R. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 6, 633–643 (2006).
Bernelot Moens, S. J. et al. Impact of the B cell growth factor APRIL on the qualitative and immunological characteristics of atherosclerotic plaques. PLoS One 11, e0164690 (2016).
Haselmayer, P., Vigolo, M., Nys, J., Schneider, P. & Hess, H. A mouse model of systemic lupus erythematosus responds better to soluble TACI than to soluble BAFFR, correlating with depletion of plasma cells. Eur. J. Immunol. 47, 1075–1085 (2017).
Tsiantoulas, D. et al. B cell-activating factor neutralization aggravates atherosclerosis. Circulation 138, 2263–2273 (2018).
Tsiantoulas, D. et al. Increased plasma IgE accelerate atherosclerosis in secreted IgM deficiency. Circ. Res. 120, 78–84 (2017).
Dishman, A. F. et al. Evolution of fold switching in a metamorphic protein. Science 371, 86–90 (2021).
Schillinger, M. et al. Inflammation and Carotid Artery—Risk for Atherosclerosis Study (ICARAS). Circulation 111, 2203–2209 (2005).
Winkelmann, B. R. et al. Rationale and design of the LURIC study—a resource for functional genomics, pharmacogenomics and long-term prognosis of cardiovascular disease. Pharmacogenomics 2 (Suppl 1), S1–S73 (2001).
Puymirat, E. et al. Acute myocardial infarction: changes in patient characteristics, management, and 6-month outcomes over a period of 20 years in the FAST-MI program (French Registry of Acute ST-Elevation or Non-ST-Elevation Myocardial Infarction) 1995 to 2015. Circulation 136, 1908–1919 (2017).
Kowalczyk-Quintas, C. et al. Generation and characterization of function-blocking anti-ectodysplasin A (EDA) monoclonal antibodies that induce ectodermal dysplasia. J. Biol. Chem. 289, 4273–4285 (2014).
Chou, M. Y. et al. Oxidation-specific epitopes are dominant targets of innate natural antibodies in mice and humans. J. Clin. Invest. 119, 1335–1349 (2009).
Kijani, S., Vázquez, A. M., Levin, M., Borén, J. & Fogelstrand, P. Intimal hyperplasia induced by vascular intervention causes lipoprotein retention and accelerated atherosclerosis. Physiol. Rep. 5, e13334 (2017).
Tom, R., Bisson, L. & Durocher, Y. Transfection of HEK293-EBNA1 cells in suspension with linear PEI for production of recombinant proteins. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot4977 (2008).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bossen, C. et al. TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood 111, 1004–1012 (2008).
Kowalczyk-Quintas, C. et al. Inhibition of membrane-bound BAFF by the anti-BAFF antibody Belimumab. Front. Immunol. 9, 2698 (2018).
Schneider, P., Willen, L. & Smulski, C. R. Tools and techniques to study ligand-receptor interactions and receptor activation by TNF superfamily members. Methods Enzymol. 545, 103–125 (2014).
Manza, L. L., Stamer, S. L., Ham, A.-J. L., Codreanu, S. G. & Liebler, D. C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 (2005).
Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Olsen, J. V. et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 (2005).
Hellings, W. E., Moll, F. L., de Kleijn, D. P. & Pasterkamp, G. 10-years experience with the Athero-Express study. Cardiovasc. Diagn. Ther. 2, 63–73 (2012).
Mayer, F. J. et al. Combined effects of inflammatory status and carotid atherosclerosis: a 12-year follow-up study. Stroke 47, 2952–2958 (2016).
Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Acknowledgements
We thank A. Fabry and T. Wenko for help with the in vivo experimental studies, and C. Friedl for help with confocal microscopy. This work was supported by grants from the Austrian Science Fund (SFB F54), the European Union (FP7 VIA) and the Leducq Foundation (TNE-20CVD03) to C.J.B., and by grants from the European Research Area Network on Cardiovascular Diseases (I4647) and the British Heart Foundation (RCAG/917) to D.T. D.T. is also supported by the Austrian Science Fund (I4963). P.S. is supported by the Swiss National Science Foundation (31003A_176256, 310030E_197000). Z.M. is supported by the British Heart Foundation (RCAM/104, RCAM-659, RRCAM.163), the British Heart Foundation Center for Research Excellence (RE/18/1/34212), the NIHR Cambridge Biomedical Research Centre (RG85315), the European Union (FP7 VIA; RCAG/430) and the European Research Council (ERC). T.H. is supported by a NWO Veni grant (91619012).
Author information
Authors and Affiliations
Contributions
D.T. conceived and designed the study, performed most of the experiments, analysed and interpreted data, and wrote the manuscript. M.E. and P.S. generated materials, performed experiments to characterize nc-APRIL and interpreted data. G.O., L.E., S.K., L.W., T.A., T.H., M.G.K., M.O.-K., L.G., F.P., J.E.M. and P.F. aided in mouse studies and provided technical assistance with the experiments. M.C. aided in immunofluorescence analyses. D.S. aided in mouse studies. J.L. and H.F.J. provided the RNA-seq data for mouse VSMCs. A.M. and J.W. performed the mass-spectrometry analysis and analysed the data. F.J.M. and F.F. performed statistical analysis of the ICARAS and LURIC clinical data, respectively. O.D. and J.B. provided reagents and technical expertise with the experiments, and critically revised the manuscript. M.H. was involved in the analysis of human samples. T.S and N.D. provided the samples from the FAST-MI clinical study and performed the statistical analysis of the data. H.S., W.M. and Z.M. provided the samples from the LURIC clinical study and measured nc-APRIL levels. G.P. provided materials. H.H. provided materials and critically revised the manuscript. Z.M. and P.S. contributed to study design, interpreted data and critically revised the manuscript. C.J.B. designed the study, interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.T., C.J.B., P.S. and M.E. are named inventors on a patent application (EP20217536.0; pending) to exploit c-APRIL and nc-APRIL for diagnostic and therapeutic purposes in cardiovascular disease that has been filed by the Medical University of Vienna (Austria) and CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences (Austria). O.D. is an employee of Adipogen Life Sciences, which provided some reagents used in this study.
Additional information
Peer review information Nature thanks Peter Libby, William Sessa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
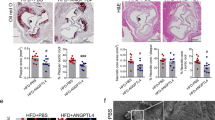
Extended Data Fig. 1 APRIL deficiency does not alter the plaque smooth muscle cell and collagen content, or the numbers of circulating monocytes, B or T lymphocytes in Ldlr−/− mice.
Ldlr−/−Tnfsf13+/+ or Ldlr−/−Tnfsf13−/−mice were fed an atherogenic diet for 10 weeks. a, Whole body weight, plasma triglyceride levels and en face lesion size (n = 10 Ldlr−/−Tnfsf13+/+ mice, n = 12 Ldlr−/−Tnfsf13−/− mice). b–d, Representative photomicrographs of DAPI- (b), α-SMA- (c) and Sirius Red-stained lesions (d) in the aortic origin (left) and dot plots (right) showing the averaged acellular (b; n = 9 Ldlr−/−Tnfsf13+/+ mice, n = 12 Ldlr−/−Tnfsf13−/− mice, P = 0.036), α-SMA-positive (c, n = 9 Ldlr−/−Tnfsf13+/+ mice, n = 10 Ldlr−/−Tnfsf13−/− mice) and collagen-positive area normalized to total lesion size (d; n = 10 Ldlr−/−Tnfsf13+/+ mice, n = 12 Ldlr−/−Tnfsf13−/− mice). e–g, Representative flow cytometry dot plots (top) and quantification (bottom) of the absolute numbers of splenic CD3+, CD4+ (defined as CD3+CD4+CD8−) and CD8+ T cells (defined as CD3+CD8+CD4−) (e), frequencies of peritoneal B-1a (defined as B220lowCD11bintCD5+), B-1b (defined as B220lowCD11bintCD5−) and CD23+ B-2 (defined as B220highCD11b−CD5−CD23+) cells (f) and the frequencies of circulating Ly6Chigh, Ly6Cint and Ly6Clow monocytes (g) in peripheral blood. All results show mean (two-tailed unpaired Student’s t-test). Scale bars, 200 μm.
Extended Data Fig. 2 BCMA is dispensable for atherosclerosis development.
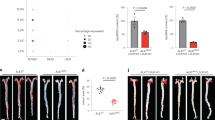
Lethally irradiated Ldlr−/− mice were injected with bone marrow from Bcma+/+ (hem-Bcma+/+) or Bcma−/− donors (hem-Bcma−/−) and were fed an atherogenic diet for 10 weeks. a, Bcma mRNA in the spleen (n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). b, Representative photomicrographs of H&E-stained aortic root lesions (left) and average lesion size in the aortic origin (right) expressed as μm2 per section (n = 11 hem-Bcma+/+, n = 12 hem-Bcma−/− mice). c, Whole body weight, plasma triglyceride levels and en face lesion size (n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). d, Total plasma cholesterol (n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). e, Absolute numbers of FO/T2, MZ, CD21+CD23− (P = 0.028), T1 (P = 0.023), NF (P = 0.031) and B-1 B cells (n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). f, Absolute numbers of CD3+, CD4+ and CD8+ T cells (n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). g, h, Frequencies of peritoneal B-1a (P = 0.003), B-1b (P = 0.021) and CD23+ B-2 cells (P = 0.0003) (g) and total IgM (P = 0.002), IgG1, IgG2b (P = 0.002), IgG2c (P = 0.001), IgG3 (P = 0.032) and IgA plasma antibody titers (h; n = 11 hem-Bcma+/+, n = 13 hem-Bcma−/− mice). All results show mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (two-tailed Mann–Whitney U-test or two-tailed unpaired Student’s t-test). Scale bar, 200 μm.
Extended Data Fig. 3 APRIL is produced by mouse and human VSMCs.
a, TNFSF13 gene expression in human tissues in the Genotype-Tissue Expression (GTEx) project36. The GTEx project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data described in this manuscript were obtained from the GTEx Portal on 21 January 2021 and dbGaP accession number phs000424.v8.p2. The results show median (aorta: median = 55.05, n = 432; coronary artery: median = 40.7, n = 240). b, c, Bulk RNA-seq analysis of VSMCs from the aortic arch (AA) and descending thoracic aorta (DT) (b; n = 3–5 mice) (GSE117963) and from mouse primary VSMCs that were stored in Trizol after isolation or had been cultured for 4–5 passages until the analysis (c; GSE17858). TNFSF13, MYH11 and KI67 gene expression are depicted. d, TNFSF13 and IL6 gene expression by human umbilical artery smooth muscle cells that were stimulated in quadruplicate with recombinant human TNF, native human LDL or human oxLDL (TNF stimulation is representative of three independent experiments; P = 0.003). Results show mean ± s.e.m. **P < 0.01 (one-way ANOVA and Tukey’s test).
Extended Data Fig. 4 APRIL binds HSPG2.
a, Representative photomicrographs (left) and quantification (right) of HUVECs incubated with either human or mouse Flag–APRIL in the presence or absence of heparin and stained with the anti-Flag M2 antibody conjugated to FITC and analysed by confocal microscopy. b, Flow cytometry analysis of HUVECs incubated with Flag–APRIL or flag-tagged bacterial alkaline phosphatase (Flag–BAP) and stained with the anti-Flag M2 antibody. c, Identification of protein binding partners of APRIL in HUVEC culture by performing a pull-down assay with agarose beads coupled to the anti-Flag M2 antibody followed by MS analysis. d, APRIL binding to coated HSPGs from mouse basement membrane quantified by ELISA determined in triplicate (****P < 0.0001, two-tailed unpaired Student t test). Data shown are representative of at least two independent experiments (a, b, d). e, f, Photomicrographs of mouse carotid artery sections incubated with mouse multimeric Flag–APRIL and stained with either an anti-Flag antibody conjugated to PE or with an anti-mouse APRIL biotinylated antibody (2C8) and streptavidin conjugated to PE (e; scale bar, 75 μm, data derived from two independent experiments) or with mouse multimeric Flag–APRIL in the presence or absence of heparin and stained with an anti-APRIL biotinylated antibody (2C8) or with an anti-HSPG2 or only secondary antibody (only 2nd) (f; scale bar, 75 μm, data derived from one experiment). g, Quantitative surface plasmon resonance (Biacore) analysis of the affinity of soluble human Fc–APRIL (total), human canonical Fc–APRIL (human Fc–c-APRIL), human non-canonical Fc–APRIL (human Fc–nc-APRIL), mouse canonical Fc–APRIL (mouse Fc–c-APRIL), mouse non-canonical Fc–APRIL (mouse Fc–nc-APRIL) and negative controls EDAR–Fc and human Fas–Fc to biotinylated heparin coupled to streptavidin Sensor Chip A (n = 3 independent experiments). All results show mean ± s.e.m. IntDen, integrated density.
Extended Data Fig. 5 Anti-APRIL antibodies 108, 2C8 and Apry-1-1 are specific for mouse APRIL.
a, Coomassie blue analyses of anti-mAPRIL mAb 108 and 2C8 under reducing conditions. b, Isotyping of the Fc portions of anti-mAPRIL 108, Apry-1-1 and 2C8. Purified antibodies coated on an ELISA plate were revealed with peroxidase-conjugated antibodies against different isotypes. c, Inhibitory activity of 108 and Apry-1-1 compared to that of TACI-Fc on human and mouse APRIL. Flag–human APRIL and two splice variants of Flag–mouse APRIL (±Ala112) were titrated on BCMA–Fas reporter cells in the presence of a fixed, non-saturating concentration of 108, Apry-1-1 or TACI-Fc. The data show that 108 and Apry-1-1 inhibit both splice variants of mAPRIL at roughly stoichiometric ratios, but do not cross-react with human APRIL. d, ELISA for 2C8 binding to mouse APRIL. Binding of 2C8 to plates coated with human Fc–mouse APRIL was evaluated with a peroxidase-coupled anti-mouse antibody. a, b, Data are representative of two independent experiments.
Extended Data Fig. 6 APRIL competes for binding of LDL to proteoglycans.
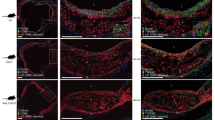
a, Representative photomicrographs (left) and quantification (right) of anti-ApoB antibody binding to mouse carotid artery sections incubated with human native LDL in the presence or absence of mouse multimeric Flag–APRIL analysed by both confocal and epifluorescence microscopy (without APRIL, n = 7; with APRIL, n = 9; P = 0.0004). b, The amount of bound human LDL (triplicate; quantified by flow cytometry) on the surface of HEK293 wild-type cells in the presence of different amounts of human recombinant APRIL. c, Representative photomicrographs of ApoB-stained lesions in the aortic origin (left) and ApoB-positive area normalized to DAPI+ lesion area (right) of Ldlr−/−Tnfsf13+/+ or Ldlr−/−Tnfsf13−/−mice that were fed an atherogenic diet for 10 weeks (n = 8 Ldlr−/−Tnfsf13+/+ mice, n = 12 Ldlr−/−Tnfsf13−/− mice; P = 0.035). Data shown are pooled from four independent experiments with seven to nine sections per group (a), representative of three independent experiments (b). All results show mean ± s.e.m. *P < 0.05, ***P < 0.001 (two-tailed unpaired Student’s t-test). Scale bars, 20 μm (a) and 200 μm (c).
Extended Data Fig. 7 Treatment with the blocking anti-APRIL antibody Apry1-1 does not alter B cells, IgM and plasma lipid levels in Apoe−/−mice.
Apoe−/− mice were treated biweekly for 10 weeks with a mixture of anti- mouse APRIL antibody (Apry-1-1) and control-Ig (anti-APRIL group), or TACI-Ig and isotype IgG2b (TACI-Ig group), or isotype IgG2b and control-Ig (control group) and were fed an atherogenic diet for the last 8 weeks of the study. a–f, Dot plots show the numbers of total splenic B cells (a), follicular (FO) B cells (b), marginal zone (MZ) B cells (c), and frequencies of peritoneal B-1a (d), peritoneal B-1b (e) and total IgM antibody levels (f) in plasma. g, Wild-type mice were injected intraperitoneally with either 1 μg mouse multimeric Flag–APRIL or a mixture of 1 μg Flag–APRIL and 10 μg anti-mouse APRIL antibody (Apry-1-1). The amount of Flag–APRIL in plasma was measured by ELISA one, three and six hours after the injection (n = 4 mice Flag–APRIL, n = 5 mice Flag–APRIL + anti-APRIL (Apry1-1)). h, Whole body weight, plasma triglyceride and cholesterol levels. a–f, h, All results show mean ± s.e.m.; (n = 10 Apoe−/− control, n = 12 Apoe−/− TACI-Ig, n = 10 Apoe−/− anti-APRIL). ***P < 0.001, ****P < 0.0001 (one-way ANOVA and Newman–Keuls test).
Extended Data Fig. 8 Epitope mapping of anti-human APRIL antibodies, and native canonical and non-canonical APRIL differ in size but are produced by the same gene.
a–g, Epitope mapping of anti-human APRIL antibodies. a, The epitopes recognized by Aprily1, 2, 3, 5 and 10 were mapped by western blot of truncated APRIL proteins. b–d, Aprily5 and Aprily1 or Aprily2 recognize distinct epitopes (d), whereas Aprily3 and Aprily10 recognize epitopes distinct from those of Aprily1, Aprily2 and Aprily5 (e, f). g, Expression of all constructs was validated by western blot with anti-Fc antibody. h, i, Human serum was depleted of APRIL using the anti-human APRIL antibodies Aprily1, Aprily2, Aprily3, Aprily5, Aprily6, Aprily8, Aprily9, Aprily10, Mahya-1, 110.6, the biological atacicept (TACI-Ig: a recombinant fusion protein of the receptor TACI and the Fc region of Ig, that binds to APRIL) or the negative control EctoD1, and then analysed with a c-APRIL-specific (h; ELISA 1) or an nc-APRIL-specific ELISA (i; ELISA 2). Data are derived from one experiment in this format. j–n, Native canonical and non-canonical APRIL differ in size. j, Flag–human APRIL (from c-APRIL ELISA 1 standards) was depleted on TACI–Fc (or TNFR2–Fc as control) and/or on Aprily2 (or mIgG1 as control). APRIL was then detected by c-APRIL-specific (top) or nc-APRIL-specific (bottom) ELISA. k, l, Flag–human APRIL (from APRIL ELISA 1 standards) was depleted on immobilized TACI–Fc or on Aprily2, and the flow-through was then size-fractionated by size exclusion chromatography (SEC) and detected in fractions by c-APRIL-specific (k; ELISA 1) or nc-APRIL-specific (l; ELISA 2) ELISA. TACI–Fc and Aprily2 beads used for depletion were then acid-eluted. m, n, The neutralized eluate was size-fractionated, and APRIL in fractions was detected with c-APRIL-specific (m) or nc-APRIL-specific (n) ELISA. These results indicate that Flag–c-APRIL has the size of a 3-mer, whereas nc-APRIL is much larger. o, p, Canonical and non-canonical APRIL are produced by the same TNFSF13 gene locus. The TNFSF13 gene (which encodes APRIL) was inactivated in human macrophage cell line U937 by CRISPR–Cas9 technology. As a control, the TNFSF13B gene (which encodes BAFF) was also deleted. APRIL in supernatants was measured with a c-APRIL-specific (o) and an nc-APRIL-specific (p) ELISA. 105, 110, 301 and 302 depict different clones.
Extended Data Fig. 9 LC–MS-based parallel reaction monitoring (PRM) analysis of tryptic digest of purified human canonical or non-canonical Fc–APRIL.
a–c, Raw data were analysed using Skyline software and extracted product ion chromatograms (XICs) are shown either in the form of peaks (top) or total sum of integrated product ion areas (bottom) for the three selected peptides EEQYNSTYR (Fc part) (a), LNLSPHGTFLGFVK (tryptic C terminus APRIL) (b) and LNLSPHGTFLGFVKL (miscleaved tryptic C terminus APRIL) (c). MS2 fragment ion spectra for the selected peptide precursor ions are illustrated at bottom right. Although the peptide shown in a is representative for comparable injection amounts of canonical versus non-canonical Fc–APRIL, the C-terminal miscleaved full tryptic peptide shown in c is undetectable in non-canonical APRIL. Relative abundances are given in arbitrary units. Right, FASTA sequence of Fc–APRIL with selected tryptic peptide sequences highlighted in blue or red. Note the different scales in b (109) and c (106). d, Structure of human c-APRIL highlighting the importance of the C terminus for the folding of the different forms (canonical and non-canonical) of APRIL. The representation based on protein data bank accession number 1XU1 highlights the last two C-terminal amino acids (Lys232, Leu233). The N-terminal amino acid of the TNF homology domain (His98) and Asp142 are also shown. All of these residues are conserved in mouse APRIL and human APRIL, although the sequence surrounding Asp142 is different in mouse and human. The C-terminal carboxylic group of Leu233 is very close to His98 of the same monomer (3.6 Å, 4.1 Å and 3.4 Å in the three monomers) and also very close to His98 of the neighbouring monomer (4.3 Å, 3.8 Å and 3.8 Å). Thus, His98 and the carboxylic group or Leu233 seem to form a ring of six salt bridges at the top surface of APRIL. In addition, Lys232 contacts Asp142 (4.3 Å, 4.3 Å and 5.7 Å in the three mouse APRIL monomers), and is only 3.2 Å from Asp142 in human APRIL (PDB accession number 4ZCH).
Supplementary information
Supplementary Figure 1
Quantified purified nc-APRIL and c-APRIL used as standards in nc-APRIL specific and c-APRIL specific ELISA, respectively.
Supplementary Table 1
Contains data referring to Extended Data Figure 9a-c.
Supplementary Table 2
Contains data referring to Extended Data Table 1a, b.
Supplementary Table 3
Contains data referring to Extended Data Table 1c.
Rights and permissions
About this article
Cite this article
Tsiantoulas, D., Eslami, M., Obermayer, G. et al. APRIL limits atherosclerosis by binding to heparan sulfate proteoglycans. Nature 597, 92–96 (2021). https://doi.org/10.1038/s41586-021-03818-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03818-3
This article is cited by
-
Dysregulated cellular metabolism in atherosclerosis: mediators and therapeutic opportunities
Nature Metabolism (2024)
-
IFNγ binding to extracellular matrix prevents fatal systemic toxicity
Nature Immunology (2023)
-
Structure of the human heparan sulfate polymerase complex EXT1-EXT2
Nature Communications (2022)
-
Therapeutic strategies targeting inflammation and immunity in atherosclerosis: how to proceed?
Nature Reviews Cardiology (2022)
-
The why and how of adaptive immune responses in ischemic cardiovascular disease
Nature Cardiovascular Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.