Abstract
Reproductive longevity is essential for fertility and influences healthy ageing in women1,2, but insights into its underlying biological mechanisms and treatments to preserve it are limited. Here we identify 290 genetic determinants of ovarian ageing, assessed using normal variation in age at natural menopause (ANM) in about 200,000 women of European ancestry. These common alleles were associated with clinical extremes of ANM; women in the top 1% of genetic susceptibility have an equivalent risk of premature ovarian insufficiency to those carrying monogenic FMR1 premutations3. The identified loci implicate a broad range of DNA damage response (DDR) processes and include loss-of-function variants in key DDR-associated genes. Integration with experimental models demonstrates that these DDR processes act across the life-course to shape the ovarian reserve and its rate of depletion. Furthermore, we demonstrate that experimental manipulation of DDR pathways highlighted by human genetics increases fertility and extends reproductive life in mice. Causal inference analyses using the identified genetic variants indicate that extending reproductive life in women improves bone health and reduces risk of type 2 diabetes, but increases the risk of hormone-sensitive cancers. These findings provide insight into the mechanisms that govern ovarian ageing, when they act, and how they might be targeted by therapeutic approaches to extend fertility and prevent disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Full genome-wide association summary statistics for the discovery meta-analysis are available from the ReproGen website (www.reprogen.org). The MII oocyte dataset is available from the European Genome-Phenome Archive under accession EGAS00001004947 (https://ega-archive.org/studies/EGAS00001004947). Access to these data is granted in accordance with the ethics permissions under which the data were collected from participants and under appropriate GDPR compliant data processing agreements. Data from the following sites were used: SMR (https://cnsgenomics.com/software/smr/#eQTLsummarydata); Tabula Muris (https://tabula-muris.ds.czbiohub.org/); LDSC-SEG (https://github.com/bulik/ldsc/wiki/Cell-type-specific-analyses); RNA-seq samples (https://www.ebi.ac.uk/ena); human oocyte expression analyses: Gene Expression Omnibus GSE86146, GSE107746. Source data are provided with this paper.
References
Lambalk, C. B., van Disseldorp, J., de Koning, C. H. & Broekmans, F. J. Testing ovarian reserve to predict age at menopause. Maturitas 63, 280–291 (2009).
Collaborative Group on Hormonal Factors in Breast Cancer. Type and timing of menopausal hormone therapy and breast cancer risk: individual participant meta-analysis of the worldwide epidemiological evidence. Lancet 394, 1159–1168 (2019).
Murray, A. et al. Population-based estimates of the prevalence of FMR1 expansion mutations in women with early menopause and primary ovarian insufficiency. Genet. Med. 16, 19–24 (2014).
Christensen, K., Doblhammer, G., Rau, R. & Vaupel, J. W. Ageing populations: the challenges ahead. Lancet 374, 1196–1208 (2009).
InterLACE Study Team. Variations in reproductive events across life: a pooled analysis of data from 505 147 women across 10 countries. Hum. Reprod. 34, 881–893 (2019).
Gruhn, J. R. et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 365, 1466–1469 (2019).
Donnez, J. & Dolmans, M.-M. Fertility preservation in women. N. Engl. J. Med. 377, 1657–1665 (2017).
Yding Andersen, C., Mamsen, L. S. & Kristensen, S. G. Fertility preservation: freezing of ovarian tissue and clinical opportunities. Reproduction 158, F27–F34 (2019).
Argyle, C. E., Harper, J. C. & Davies, M. C. Oocyte cryopreservation: where are we now? Hum. Reprod. Update 22, 440–449 (2016).
Stolk, L. et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 44, 260–268 (2012).
Venturella, R. et al. The genetics of non-syndromic primary ovarian insufficiency: a systematic review. Int. J. Fertil. Steril. 13, 161–168 (2019).
Titus, S. et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci. Transl. Med. 5, 172ra21 (2013).
Day, F. R. et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat. Genet. 47, 1294–1303 (2015).
Horikoshi, M. et al. Elucidating the genetic architecture of reproductive ageing in the Japanese population. Nat. Commun. 9, 1977 (2018).
Caburet, S. et al. Homozygous hypomorphic BRCA2 variant in primary ovarian insufficiency without cancer or Fanconi anaemia trait. J. Med. Genet. 58,125–134 (2020).
Thompson, D. J. et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature 575, 652–657 (2019).
Day, F. R. et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 49, 834–841 (2017).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Reddy, P. et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science 319, 611–613 (2008).
Suh, E.-K. et al. p63 protects the female germ line during meiotic arrest. Nature 444, 624–628 (2006).
Deutsch, G. B. et al. DNA damage in oocytes induces a switch of the quality control factor TAp63α from dimer to tetramer. Cell 144, 566–576 (2011).
Tuppi, M. et al. Oocyte DNA damage quality control requires consecutive interplay of CHK2 and CK1 to activate p63. Nat. Struct. Mol. Biol. 25, 261–269 (2018).
Rinaldi, V. D., Bloom, J. C. & Schimenti, J. C. Oocyte elimination through DNA damage signaling from CHK1/CHK2 to p53 and p63. Genetics 215, 373–378 (2020).
Aiken, C. E., Tarry-Adkins, J. L., Penfold, N. C., Dearden, L. & Ozanne, S. E. Decreased ovarian reserve, dysregulation of mitochondrial biogenesis, and increased lipid peroxidation in female mouse offspring exposed to an obesogenic maternal diet. FASEB J. 30, 1548–1556 (2016).
Pittman, D. L. et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705 (1998).
Nakajo, N. et al. Absence of Wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev. 14, 328–338 (2000).
Ruth, K. S. et al. Events in early life are associated with female reproductive ageing: a UK Biobank study. Sci. Rep. 6, 24710 (2016).
Pan, Y. et al. A heterozygous hypomorphic mutation of Fanca causes impaired follicle development and subfertility in female mice. Mol. Genet. Genomics 296, 103–112 (2021).
Bakker, S. T. et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 18, 3484–3495 (2009).
Caburet, S. et al. Mutant cohesin in premature ovarian failure. N. Engl. J. Med. 370, 943–949 (2014).
Bolcun-Filas, E., Rinaldi, V. D., White, M. E. & Schimenti, J. C. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 343, 533–536 (2014).
Adhikari, D. et al. Inhibitory phosphorylation of Cdk1 mediates prolonged prophase I arrest in female germ cells and is essential for female reproductive lifespan. Cell Res. 26, 1212–1225 (2016).
Rinaldi, V. D., Bolcun-Filas, E., Kogo, H., Kurahashi, H. & Schimenti, J. C. The DNA damage checkpoint eliminates mouse oocytes with chromosome synapsis failure. Mol. Cell 67, 1026–1036.e2 (2017).
Tharp, M. E., Malki, S. & Bortvin, A. Maximizing the ovarian reserve in mice by evading LINE-1 genotoxicity. Nat. Commun. 11, 330 (2020).
Liu, Q. et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000).
Abe, H. et al. CHEK1 coordinates DNA damage signaling and meiotic progression in the male germline of mice. Hum. Mol. Genet. 27, 1136–1149 (2018).
Chen, L. et al. Checkpoint kinase 1 is essential for meiotic cell cycle regulation in mouse oocytes. Cell Cycle 11, 1948–1955 (2012).
Pacheco, S. et al. ATR is required to complete meiotic recombination in mice. Nat. Commun. 9, 2622 (2018).
Pacheco, S., Maldonado-Linares, A., Garcia-Caldés, M. & Roig, I. ATR function is indispensable to allow proper mammalian follicle development. Chromosoma 128, 489–500 (2019).
López-Contreras, A. J., Gutierrez-Martinez, P., Specks, J., Rodrigo-Perez, S. & Fernandez-Capetillo, O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 209, 455–461 (2012).
Salpeter, S. R. et al. Meta-analysis: effect of hormone-replacement therapy on components of the metabolic syndrome in postmenopausal women. Diabetes Obes. Metab. 8, 538–554 (2006).
Manson, J. E. et al. Menopausal hormone therapy and health outcomes during the intervention and extended poststopping phases of the Women’s Health Initiative randomized trials. J. Am. Med. Assoc. 310, 1353–1368 (2013).
Ruth, K. S. et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 26, 252–258 (2020).
Dam, V. et al. Association of menopausal characteristics and risk of coronary heart disease: a pan-European case-cohort analysis. Int. J. Epidemiol. 48, 1275–1285 (2019).
de Kat, A. C. et al. Unraveling the associations of age and menopause with cardiovascular risk factors in a large population-based study. BMC Med. 15, 2 (2017).
Atsma, F., Bartelink, M.-L. E. L., Grobbee, D. E. & van der Schouw, Y. T. Postmenopausal status and early menopause as independent risk factors for cardiovascular disease: a meta-analysis. Menopause 13, 265–279 (2006).
Ambikairajah, A., Walsh, E. & Cherbuin, N. Lipid profile differences during menopause: a review with meta-analysis. Menopause 26, 1327–1333 (2019).
Pike, C. J. Sex and the development of Alzheimer’s disease. J. Neurosci. Res. 95, 671–680 (2017).
Zhu, D. et al. Body mass index and age at natural menopause: an international pooled analysis of 11 prospective studies. Eur. J. Epidemiol. 33, 699–710 (2018).
Shadyab, A. H. et al. Ages at menarche and menopause and reproductive lifespan as predictors of exceptional longevity in women: the Women’s Health Initiative. Menopause 24, 35–44 (2017).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Tyrrell, J. et al. Using genetics to understand the causal influence of higher BMI on depression. Int. J. Epidemiol. 48, 834–848 (2019).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Terao, C. et al. Chromosomal alterations among age-related haematopoietic clones in Japan. Nature 584, 130–135 (2020).
Wood, A. R. et al. Variants in the FTO and CDKAL1 loci have recessive effects on risk of obesity and type 2 diabetes, respectively. Diabetologia 59, 1214–1221 (2016).
Szustakowski, J. D. et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. https://doi.org/10.1038/s41588-021-00885-0 (2021).
McLaren, W. et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Zhou, W. et al. Scalable generalized linear mixed model for region-based association tests in large biobanks and cohorts. Nat. Genet. 52, 634–639 (2020).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Võsa, U. et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. Preprint at https://doi.org/10.1101/447367 (2018).
Pers, T. H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Fehrmann, R. S. N. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 47, 115–125 (2015).
Segrè, A. V., Groop, L., Mootha, V. K., Daly, M. J. & Altshuler, D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput. Biol. 11, e1004219 (2015).
Lamparter, D., Marbach, D., Rueedi, R., Kutalik, Z. & Bergmann, S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLOS Comput. Biol. 12, e1004714 (2016).
Deelen, P. et al. Improving the diagnostic yield of exome- sequencing by predicting gene-phenotype associations using large-scale gene expression analysis. Nat. Commun. 10, 2837 (2019).
Knijnenburg, T. A. et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254.e6 (2018).
Pearl, L. H., Schierz, A. C., Ward, S. E., Al-Lazikani, B. & Pearl, F. M. G. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 15, 166–180 (2015).
Álvarez-Quilón, A. et al. Endogenous DNA 3′ blocks are vulnerabilities for BRCA1 and BRCA2 deficiency and are reversed by the APE2 nuclease. Mol. Cell 78, 1152–1165.e8 (2020).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Bowden, J. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the radial plot and radial regression. Int. J. Epidemiol. 47, 2100 (2018).
Samuelsson, A.-M. et al. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertension 51, 383–392 (2008).
Sankar, A. et al. KDM4A regulates the maternal-to-zygotic transition by protecting broad H3K4me3 domains from H3K9me3 invasion in oocytes. Nat. Cell Biol. 22, 380–388 (2020).
Li, L. et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858–873.e4 (2017).
Zhang, Y. et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol. Cell 72, 1021–1034.e4 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Chen, S. et al. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 18, 80 (2017).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protocols 11, 1650–1667 (2016).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Takai, H. et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21, 5195–5205 (2002).
Lewandoski, M., Wassarman, K. M. & Martin, G. R. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148–151 (1997).
Lam, M. H., Liu, Q., Elledge, S. J. & Rosen, J. M. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell 6, 45–59 (2004).
Takeo, T. & Nakagata, N. Superovulation using the combined administration of inhibin antiserum and equine chorionic gonadotropin increases the number of ovulated oocytes in C57BL/6 female mice. PLoS One 10, e0128330 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
This research was conducted using the UK Biobank resource under application numbers 871 (Exeter) and 9797 (Cambridge). Full individual study acknowledgements can be found in the Supplementary Information. The authors wish to dedicate this work to the memory of Professor P. Solc.
Author information
Authors and Affiliations
Consortia
Contributions
All authors reviewed the original and revised manuscripts. Leads on manuscript writing: K.S.R., F.R.D., E.R.H., Anna Murray, I. Roig, J.R.B.P. Central statistical genetics analysis team: K.S.R., F.R.D., Anna Murray, J.R.B.P. Animal model working group: J.H., A.M.-M., C.E.A., L.K., H.A., J.L.T., J.M.G., S.T., E.P.T.H., M.F., Y.H., A.S., A. Pujol, A.J.L., J.A.D., S.E.O., S.H.N., P. Solc, E.R.H., I. Roig. Human oocyte expression working group: A. Azad, V.S., R.B., K.W.O., M.K.H., M.L.G., C.Y., E.R.H. Sample collection, genotyping, phenotyping and individual study analysis: K.S.R., F.R.D., D.J.T., P.F., A. Claringbould, O.B.B., P. Sulem, R.G.W., C.T., M.H., K.L., N.O., P.N.T., P.A., S. Stankovic, P.R.H.J.T., T.U.A., B.Z.A., E.N., I.L.A., A.M.A., K.J.A., A. Augustinsson, S. Bandinelli, C.M.B., R.N.B., H.B., M.W.B., S. Benonisdottir, S. Bergmann, M.B., E.B., S.E.B., M.K.B., D.I.B., N.B., J.A.B., L.B., J.E.B., A. Campbell, H.C., J.E.C., E.C., S.J.C., G.C., M.C., T.C., F.J.C., A. Cox, L.C., S.S.C., F.C., K.C., G.D., E.J.C.N.D, R.D., I.D., E.W.D., J.D., A.M.D., M.D., M.E., T.E., P.A.F., J.D.F., L. Ferrucci, N.F., T.M.F., M.G.-D., M. Mezzavilla, M.G.-C., C.G., G.G.G., H.G., D.F.G., V.G., P.G., C.A.H., N.H., P.H., C. Hayward, C. He, W.H., G.H., J.L.H., J.J.H., F.H., D.H., M.A.I., R.D.J., M.D.R.J., E.M.J., P.K.J., D.K., S.L.R.K., C. Kartsonaki, R.K., C.M.K., I.K., C. Kooperberg, P.K., A.W.K., Z.K., M. La Bianca, G.L., C.L., L.J.L., J.S.E.L., D.A.L., L.L.M., J. Li, A.L., S. Lindstrom, T.L., M. Linet, Y.L., S. Liu, J. Luan, R.M., P.K.E.M., M. Mangino, A. Mannermaa, B. Marco, J. Marten, N.G.M., H.M., B.McK., S.E.M., C. Meisinger, T.M., C. Menni, A. Metspalu, L.M., R.L.M., G.W.M., D.O.M., A. Mulas, A.M.M., Alison Murray, M.A.N., A.N., R.N., T.N., D.R.N., A.F.O., H.O., J.N.P., A.V.P., N.L.P., N.P., A. Peters, U.P., P.D.P.P., O.P., E. Porcu, B.M.P., I. Rahman, G.R., H.S.R., P.M.R., S.M.R., A.R., L.M.R., F.R.R., J.R., I. Rudan, R.R., D.R., C.F.S., E.S., D.P.S., S. Sanna, E.J.S., C. Sarnowski, D. Schlessinger, M.K.S., M.J.S., K.E.S., C. Scott, S. Shekari, A.V.S., B.H.S., J.A.S., R.S., M.C.S., T.D.S., J.J.S., M.S., D. Stöckl, J.B.J.v.M., K. Strauch, U.S., A.J.S., T. Tanaka, L.R.T., A.T., U.Þ., N.J.T., D.T., M.T., M.A.T., T. Truong, J.T., A.G.U., S.U., C.M.V., V.V., U.V., P.V., H.V., Q.W., N.J.W., C.R.W., D.R.W., A.N.W., K.W., G.W., J.F.W., B.H.R.W., A.W., A.R.W., W.Z., M.Z., Z.C., L. Li, L. Franke, S. Burgess, P.D., T.H.P., K. Stefansson, J.C., Y.T.v.d.S., K.L.L., D.I.C., D.F.E., J.A.V., J.M.M., K.K.O., Anna Murray, J.R.B.P.
Corresponding authors
Ethics declarations
Competing interests
Full individual study and author disclosures can be found in the Supplementary Information.
Additional information
Peer review information Nature thanks Heng-Yu Fan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
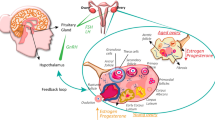
Extended Data Fig. 1 Overview of ovarian reserve and follicular activity across reproductive life.
a, Key processes involved in follicular activity from fetal development to menopause showing the numbers of oocytes at each stage. b, Summary of key biological pathways involved in follicular activity and their relationship to stage of reproductive life. Follicles, consisting of oocytes and surrounding granulosa cells, are formed in utero and maintained as resting primordial follicles in the cortex, constituting the ovarian reserve. Follicles are sequentially recruited from the ovarian reserve at a rate of several hundred per month in childhood, peaking at around 900 per month at approximately 15 years of age. Following recruitment, follicles grow by mitotic division of granulosa cells and expansion of oocyte volume for almost six months until meiosis is reinitiated at ovulation and the mature oocyte is released into the oviduct. Waves of atresia (follicle death) accompany developmental transitions and growing follicles are continuously induced to undergo cell death such that, typically, only a single follicle matures to ovulate each month. As ovarian reserve declines, the rate of follicle recruitment decreases, but the preovulatory follicles continue to produce substantial amounts of oestrogen, while other important hormones such as anti-Müllerian hormone and inhibin-B decline, leading to upregulation of the hypothalamus–pituitary gonadal axis.
Extended Data Fig. 2 Overview of performed analyses.
Analyses are separated into those intended to lead to understanding of biological mechanisms, and those that lead to insights into population genetics or epidemiology.
Extended Data Fig. 3 Consistency of effect estimates across analysis methods and strata.
a–c, Comparison of effect estimates from: a, Cox proportional hazards regression in UK Biobank with linear regression effect estimates from the overall meta-analysis (‘Effect full meta-analysis’); b, Cox proportional hazards regression in UK Biobank with linear regression effect estimates from the meta-analysis excluding UK Biobank (‘Effect 1KG+BCAC’); c, linear regression in UK Biobank with linear regression effect estimates from the meta-analysis excluding UK Biobank (‘Effect 1KG+BCAC’). d–g, Comparison of linear regression effect estimates from: d, UK Biobank GWAS versus the meta-analysis of 1000 Genomes imputed studies; e, UK Biobank GWAS versus meta-analysis of samples from the Breast Cancer Association Consortium (BCAC); f, meta-analysis of BCAC samples versus the meta-analysis of 1000 Genomes imputed studies; g, 23andMe replication analysis (rescaled) versus overall meta-analysis. HR, hazard ratio from Cox proportional hazards model; r, Pearson correlation coefficient; blue line is y = x for reference. Note: P values <1 × 10−300 are shown as 1 × 10−300.
Extended Data Fig. 4 Deviation from additive effects and distribution of estimated heritability across chromosomes.
a–d, Genome-wide significant signals showing departure from an additive model. We tested the identified signals for departure from an additive allelic model. a, rs11668344 shows no deviation from an additive allelic model, b, c, rs11670032 (b) and rs28416520 (c) show deviation from the additive allelic model and a recessive effect. d, rs75770066 shows a heterozygote effect. The mean and 95% confidence interval around the mean estimate are shown for each genotype. The expected mean ANM for the heterozygotes is the average of the mean ANM in the homozygote groups. The dashed orange line shows the effect estimate by genotype from linear regression based on an additive allelic model. Estimated ANM for each genotype was calculated as constant from regression model + number alleles × effect estimate from regression model. The dashed grey line indicates the additive effect estimate by genotype from a model adjusting for the dominance deviation effect of the heterozygote group (solid grey line). All regression models were adjusted for centre, genotyping chip and genetic principal components. dom dev, dominance deviation. e, The percentage of the total heritability explained that was attributable to each chromosome (observed heritability) is compared with the expected proportion calculated on the basis of chromosome size. The heritability of ANM was not uniformly distributed across chromosomes in proportion to their size. The X chromosome did not explain more heritability than expected given its size, but chromosome 19 explained 2.36% (1.98–2.75) of the trait variance—greater than the individual contributions of nearly all larger chromosomes (weighted average for chromosomes 1–18: 1.7%, s.e 0.2%) and about 2.5 times more than expected given its size. This was partially attributable to a single locus at 19q13 which explained about 0.75% of trait variance and where we mapped six independent signals (Supplementary Table 2). The dashed line shows the mean ratio of expected to observed heritability across all chromosomes. Chromosome size was estimated based on the number of genetic variants.
Extended Data Fig. 5 Gene co-regulation networks for age at menopause genes with MCM8 highlighted.
a, Gene co-regulation network for genes relating to age at menopause. Nodes indicate genes that are either in a cis region from the GWAS or have been prioritized by Downstreamer; edges indicate a co-regulation relationship with a z-score > 4. Co-regulation is defined as the Pearson correlation between genes in a scaled eigenvector matrix derived from a multi-tissue gene network72. cis genes are defined as genes that are within 300 kb of a GWAS top hit for age at menopause. trans genes are defined as having been prioritized by Downstreamer’s co-regulation analysis and not being within 300 kb of a GWAS top hit. Downstreamer prioritizes genes by associating the gene P value profile of the GWAS (calculated using PASCAL71) to the co-regulation profile of each protein-coding gene. Only genes for which this association passes Bonferroni significance are shown as trans genes. Teal, cis genes; dark teal, trans genes; yellow, genes with a first-degree relation to MCM8. b, Gene co-regulation network showing the genes that have a first-degree relationship with MCM8 with a z-score > 4. Edge width indicates the z-score of the co-regulation relationship. Colours as in a, with the exception of yellow, as all genes have a first-degree relation to MCM8.
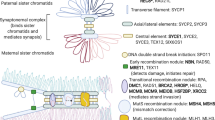
Extended Data Fig. 6 DNA damage response and repair pathways implicated in reproductive ageing in humans.
a, Consequences of replication stress annotated with genes involved that were within 300 kb of the ANM signals. b, Genes involved in downstream DNA damage response and repair pathways with those within 300 kb of an ANM signal shown in blue. A full list of genes involved in DNA damage response and apoptosis annotated with genome-wide signals for ANM is provided in Supplementary Table 19. MRN, MRN–MRE11–RAD50–NBS1 complex; RPA, replication protein A including a subunit encoded by RPA1; RFC, replication factor C including a subunit encoded by RFC1; 9-1-1, RAD9–HUS1–RAD1 complex.
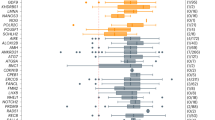
Extended Data Fig. 7 Cluster plot of expression of consensus genes identified from the genome-wide analyses in germ cells across different developmental stages.
Genes were selected from the GWAS signals, based on in silico prioritization (Supplementary Table 5). Of the 283 consensus genes highlighted by the GWAS, 258 passed QC and were available in the expression dataset. Gene expression was measured in human fetal primordial germ cells84,85 and in oocytes and granulosa cells in adult follicles (dataset generated in this study). Plot shows z-scores, calculated by subtracting the mean TPM in all samples for a gene and dividing by the s.d. GC, granulosa cell; MII, meiosis II; PGC, primordial germ cell; Wks, weeks.
Extended Data Fig. 8 Relationship between decreased ovarian reserve and gene expression.
Open bar/dot groups: control maternal diet, normal ovarian reserve. Grey bar/dot groups: obesogenic maternal diet, reduced ovarian reserve. Dots, individual observations. Bar heights and error bars: mean ± s.e.m. a, Ovarian follicular reserve in young adulthood in wild-type mice. Total follicles per mm3 ovarian tissue at 12 weeks. n = 8 biologically independent animals from different litters in each group. P = 0.0091. b, Brsk1 expression in the same mice, measured using qRT–PCR and expressed as average copy number. P = 0.0001. c, Wee1 expression in the same mice, measured using qRT–PCR and expressed as average copy number. P = 0.0256. d, Dmc1 expression in the same mice, measured using qRT–PCR and expressed as average copy number. P = 0.00001. e, Mapt expression in the same mice, measured using qRT-PCR and expressed as average copy number. P = 0.0378. All panels, two-way ANOVA after correction for multiple hypothesis testing. *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Fig. 9 Chek2 deletion increases reproductive lifespan in mouse.
a, Representative images of ovarian sections of 1.5- and 13.5-month-old wild-type (WT) and Chek2−/− mice stained with PAS-haematoxylin. Primordial follicles (inset (i)), primary follicles (inset (ii)), secondary follicle (white arrow) and antral follicle (black arrow) are shown. Scale bars, 200 μm. b–e, Number of follicles (by class and total) present in WT and Chek2−/− mouse ovaries. b, c, 1.5 months old; d, e, 13.5 months old. Numbers in parentheses show the total number of ovaries analysed. f, Serum AMH (ng ml−1) in 16–17-month-old Chek2−/− mice. Numbers in parentheses show the number of mice assessed. g–i, Gonadotrophin stimulation of 13.5-month-old females. Numbers in parentheses show: g, the number of MII oocytes retrieved per female; h, the number of MII oocytes fertilized; and i, the number of fertilized oocytes assessed for blastocyst formation. j, Litter size of WT and Chek2−/− females throughout the reproductive lifespan. Litter sizes from nine WT and five Chek2−/− females are shown. Breeding cages contained one male and one female. Generalized linear model analysis showed maternal age effect, but no effect of genotype on litter sizes. k, Image of healthy pups born to 13 month-old Chek2−/− females. b–i, Two-sample t-test and Fisher’s exact test were used to compare WT and Chek2−/− for statistical significance: *P < 0.05, **P < 0.025, ***P < 0.001. All P values are two-sided. Error bars indicate s.e.m. For boxplots: centre line, median; box limits, IQR; whiskers, 1.5 × IQR (b–g). an, antral follicle; pri, primary follicle; P0, primordial follicle; sec, secondary follicle. Mouse strain: maintained on a mixed background, C57BL/6 129Sv, accession number BRC03481 at the RIKEN Bioresource Centre.
Extended Data Fig. 10 Conditional knockout Chek1 females are infertile due to requirement for Chek1 during preimplantation embryo development.
a, Schematic of generation of conditional knockout mouse model of Chek1 (Chek1 cKO) in the female germline using Ddx4-Cre. A similar approach was used for Zp3-Cre. b, In ovarian sections stained with H&E, we found follicles, corpora lutea (CL) and oocytes which contain nuclear structures (arrowheads, magnified right panels). These findings suggest that oestrus cycles and ovulation followed by corpus luteum formation are independent of Chek1 disruption in oocytes in vivo. c, Litter size of Chek1 cKO females. Three females older than 5 weeks of age were mated with C57BL/6J males. Five independent littermate females (F/+, Tg−/Tg−; F/F, Tg−/Tg−; or F/+, Tg+/Tg−) were used as Chek1 controls (ctrl). While Chek1 control females delivered normally, Chek1 cKO females delivered no litters (Mann–Whitney test, **P = 0.0179). Thus, these results indicate that CHEK1 is essential in the female germline. d, Litter size of Chek1-cKO and control females using the Zp3-Cre during follicular growth. Three-month-old control (Chek1 F/F; Chek1 ctrl, n = 4) and conditional knockout (Chek1 F/F; Chek1 cKO with Zp3-Cre, n = 4) were consecutively mated three times with wild-type (Chek1+/+) males, and the number of live (left) and dead (right) pups was monitored. While Chek1 ctrl females delivered a normal number of live pups, Chek1 cKO females had only a reduced number of perinatally dead pups (Mann–Whitney U-test: ***P < 0.001, **P < 0.01). Numbers in parentheses show the number of litters. e, The mean number of all ovulated eggs (sum of MII oocytes and fertilized MII oocytes) per mouse with s.e.m. (Mann–Whitney U-test, P = 0.126). Each data point represents the number of eggs per mouse. Three- to five-month-old Chek1 ctrl (n = 3) and Chek1 cKO (n = 5) females were mated with wild-type (Chek1+/+) males after pMSG + hCG stimulation. The number of ovulated eggs isolated 18 h after hCG stimulation and additional 10 h cultured in vitro was scored. The number of mice is shown in parentheses. f, The proportion of fertilized MII oocytes to all ovulated eggs with a binomial confidence interval (Fisher’s exact test, *P = 0.012; 95% CI 1.9–6.0; OR, 2.62). Numbers in parentheses show the total number of analysed eggs. g, The proportion of embryos that developed to blastocysts with binomial confidence interval (Fisher’s exact test, ***P < 0.0001). Fertilized MII oocytes (zygotes) were isolated from females stimulated with pMSG + hCG 18 h after hCG administration and cultured in vitro for 96 h (approximately embryonic day (E)3.5) when development to blastocyst was scored. Data are pooled from four independent experiments. The number of embryos is shown in parentheses. h, Fertilized eggs from Chek1 ctrl (n = 18) and Chek1 cKO (n = 13) females were fixed and stained for DNA (DAPI). All fertilized eggs from both genotypes showed normal pronucleus formation. The data were pooled from two independent experiments. Asterisks, polar bodies. i, The majority of Chek1 ctrl embryos formed blastocysts (g), but Chek1 cKO embryos were arrested mainly at the 3–8-cell stages. Representative bright-field images are shown. j, Proportion of developmental stages 2 cell, 3–4 cell and 5–8 cell (Cochran–Armitage trend test, **P = 0.0073). Chek1 ctrl and Chek1 cKO zygotes were isolated from 13 Chek1 ctrl and 6 Chek1 cKO females stimulated with pMSG + hCG 18 h after hCG administration and cultured in vitro for 49 h. Embryos were fixed and stained for γH2AX by immunofluorescence. DNA was visualized by DAPI (l). k, Proportion of embryos with genome fragmentation with binomial confidence interval (Fisher’s exact test, ***P < 0.0001). Data are pooled from two independent experiments. The number of embryos is shown in parentheses. l, Chek1 ctrl and Chek1 cKO zygotes (j, k) were fixed and stained for γH2AX (magenta) by immunofluorescence. DNA (grey) was visualized by DAPI. Arrows, genome fragments; asterisks, polar bodies. These findings suggest that maternally expressed Chek1 is critical for genome integrity protection during first divisions of preimplantation embryos in mice. All P values are two sided. For boxplots: centre line, median; box limits, IQR; whiskers, 1.5 × IQR. Strains: C57BL/6-FVB mixed background for a–c (Chek1 cKO, Ddx4-Cre); C57BL6-CD1 mixed background (Chek1 cKO, Zp3-Cre) for d–l.
Extended Data Fig. 11 Extended reproductive lifespan in females carrying an extra copy of Chek1 (sChek1).
a, mRNA expression levels of Chek1 in oocytes; numbers in parentheses show the number of mice stimulated for retrieval of oocytes. b, Representative images of ovarian sections from 1.5- and 13.5-month-old wild-type (WT) and sChek1 mice stained with PAS-haematoxylin. Primordial follicles (inset (i)), primary follicles (inset (ii)), secondary follicle (white arrow) and antral follicle (black arrow) are shown. Scale bars, 200 μm. c–f, Numbers of follicles (by class and total) in WT and sChek1 littermates: c, d, 1.5 months old; e, f, 13.5 months old. Numbers in parentheses show the total number of ovaries analysed. g–j, MII oocytes retrieved in response to pMSG and hCG (g), proportion of euploid oocytes (h), proportion fertilized (i) and proportion developed to blastocysts (j) at different ages of WT and sChek1 mice. Numbers in parentheses show: g, the number of female mice stimulated for retrieval of oocytes; h, the number of oocytes assessed for aneuploidy; i, the number of MII oocytes fertilized; and j, the number of fertilized oocytes assessed for blastocyst development. k, Proportion of live births relative to transferred embryos from in vitro fertilized oocytes from aged mice (16 months); numbers in parentheses show the number of embryos transferred. l, Healthy pups born to 16-month-old sChek1 females after IVF. m, Litter sizes from F2 females or males from aged sChek1 females after IVF treatment in k, compared to females of equivalent ages that were naturally breeding. Note that for natural breeding there were two females and one male per breeding cage, whereas F2 cages contained a single male and one female. Therefore, litter sizes are an underestimate for the IVF-conceived pups. n, Litter sizes of WT and sChek1 females throughout their reproductive lifespan. Data are from six breeding cages, three for each genotype. Each breeding cage contained one WT male and two females that were either WT or sChek1. Generalized linear model analysis showed maternal age effect, but no effect of genotype on litter sizes. a–k, Two-sample t-test and Fisher’s exact test were used to compare WT and sChek1 for statistical significance: *P < 0.05, **P < 0.025, ***P < 0.001. All P-values are two sided. Error bars indicate s.e.m. For boxplots: centre line, median; box limits, IQR; whiskers, 1.5 × IQR (c–g, m). NB, natural breeding; F2-f, F2 female; F2-m, F2 male. Mouse strain: inbred from mixed background C57BL/6 129Sv.
Supplementary information
Supplementary Information
This file contains Supplementary Results, Supplementary References and Supplementary Notes (acknowledgments, sources of funding, disclosures and consortium membership).
Supplementary Tables
This file contains Supplementary Tables 1-26 and a contents page.
Rights and permissions
About this article
Cite this article
Ruth, K.S., Day, F.R., Hussain, J. et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature 596, 393–397 (2021). https://doi.org/10.1038/s41586-021-03779-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03779-7
This article is cited by
-
No evidence for a diminished ovarian reserve among patients with hypertensive disorders of pregnancy: a case control study
Journal of Ovarian Research (2024)
-
Premature ovarian insufficiency: a review on the role of tobacco smoke, its clinical harm, and treatment
Journal of Ovarian Research (2024)
-
Genetic associations of protein-coding variants in venous thromboembolism
Nature Communications (2024)
-
The impact of reproductive factors on the metabolic profile of females from menarche to menopause
Nature Communications (2024)
-
Mendelian randomization identifies circulating proteins as biomarkers for age at menarche and age at natural menopause
Communications Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.