Abstract
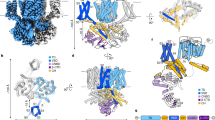
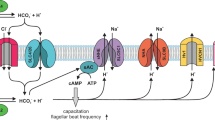
The cation channel of sperm (CatSper) is essential for sperm motility and fertility1,2. CatSper comprises the pore-forming proteins CATSPER1–4 and multiple auxiliary subunits, including CATSPERβ, γ, δ, ε, ζ, and EFCAB91,3,4,5,6,7,8,9. Here we report the cryo-electron microscopy (cryo-EM) structure of the CatSper complex isolated from mouse sperm. In the extracellular view, CATSPER1–4 conform to the conventional domain-swapped voltage-gated ion channel fold10, following a counterclockwise arrangement. The auxiliary subunits CATSPERβ, γ, δ and ε—each of which contains a single transmembrane segment and a large extracellular domain—constitute a pavilion-like structure that stabilizes the entire complex through interactions with CATSPER4, 1, 3 and 2, respectively. Our EM map reveals several previously uncharacterized components, exemplified by the organic anion transporter SLCO6C1. We name this channel–transporter ultracomplex the CatSpermasome. The assembly and organization details of the CatSpermasome presented here lay the foundation for the development of CatSpermasome-related treatments for male infertility and non-hormonal contraceptives.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM maps of the mouse CatSpermasome and the corresponding atomic coordinate have been deposited in the Electron Microscopy Data Bank and the Protein Data Bank under the accession codes EMD-31076 and 7EEB, respectively. The mass spectrometry data have been deposited in the MassIVE database (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the accession number MSV0000987325. All data analysed during this study are included in this Article and its Supplementary Information. Any other relevant data are available from the corresponding author upon reasonable request.
References
Ren, D. et al. A sperm ion channel required for sperm motility and male fertility. Nature 413, 603–609 (2001).
Ren, D. & Xia, J. Calcium signaling through CatSper channels in mammalian fertilization. Physiology 25, 165–175 (2010).
Quill, T. A., Ren, D., Clapham, D. E. & Garbers, D. L. A voltage-gated ion channel expressed specifically in spermatozoa. Proc. Natl Acad. Sci. USA 98, 12527–12531 (2001).
Lobley, A., Pierron, V., Reynolds, L., Allen, L. & Michalovich, D. Identification of human and mouse CATSPER3 and CATSPER4 genes: characterisation of a common interaction domain and evidence for expression in testis. Reprod. Biol. Endocrinol. 1, 53 (2003).
Liu, J., Xia, J., Cho, K. H., Clapham, D. E. & Ren, D. CatSperβ, a novel transmembrane protein in the CatSper channel complex. J. Biol. Chem. 282, 18945–18952 (2007).
Wang, H., Liu, J., Cho, K. H. & Ren, D. A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol. Reprod. 81, 539–544 (2009).
Chung, J. J., Navarro, B., Krapivinsky, G., Krapivinsky, L. & Clapham, D. E. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat. Commun. 2, 153 (2011).
Chung, J. J. et al. CatSperζ regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. eLife 6, e23082 (2017).
Hwang, J. Y. et al. Dual sensing of physiologic pH and calcium by EFCAB9 regulates sperm motility. Cell 177, 1480–1494 (2019).
Long, S. B., Campbell, E. B. & Mackinnon, R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908 (2005).
Whitaker, M. Calcium at fertilization and in early development. Physiol. Rev. 86, 25–88 (2006).
Okabe, M. The cell biology of mammalian fertilization. Development 140, 4471–4479 (2013).
Wang, H. F., McGoldrick, L. L. & Chung, J. J. Sperm ion channels and transporters in male fertility and infertility. Nat. Rev. Urol. 18, 46–66 (2021).
Chung, J. J. et al. Structurally distinct Ca2+ signaling domains of sperm flagella orchestrate tyrosine phosphorylation and motility. Cell 157, 808–822 (2014).
Seifert, R. et al. The CatSper channel controls chemosensation in sea urchin sperm. EMBO J. 34, 379–392 (2015).
Yoshida, M. & Yoshida, K. Sperm chemotaxis and regulation of flagellar movement by Ca2+. Mol. Hum. Reprod. 17, 457–465 (2011).
Strunker, T. et al. The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature 471, 382–386 (2011).
Lishko, P. V., Botchkina, I. L. & Kirichok, Y. Progesterone activates the principal Ca2+ channel of human sperm. Nature 471, 387–391 (2011).
Avenarius, M. R. et al. Human male infertility caused by mutations in the CATSPER1 channel protein. Am. J. Hum. Genet. 84, 505–510 (2009).
Hildebrand, M. S. et al. Genetic male infertility and mutation of CATSPER ion channels. Eur. J. Hum. Genet. 18, 1178–1184 (2010).
Jin, Z. R. et al. Roles of CatSper channels in the pathogenesis of asthenozoospermia and the therapeutic effects of acupuncture-like treatment on asthenozoospermia. Theranostics 11, 2822–2844 (2021).
Avidan, N. et al. CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur. J. Hum. Genet. 11, 497–502 (2003).
Zhang, Y. et al. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J. Med. Genet. 44, 233–240 (2007).
Luo, T. et al. A novel copy number variation in CATSPER2 causes idiopathic male infertility with normal semen parameters. Hum. Reprod. 34, 414–423 (2019).
Wang, J. X. et al. Patient with CATSPER3 mutations-related failure of sperm acrosome reaction with successful pregnancy outcome from intracytoplasmic sperm injection (ICSI). Mol. Genet. Genom. Med. 9, e1579 (2020).
Brown, S. G. et al. Homozygous in-frame deletion in CATSPERE in a man producing spermatozoa with loss of CatSper function and compromised fertilizing capacity. Hum. Reprod. 33, 1812–1816 (2018).
Quill, T. A. et al. Hyperactivated sperm motility driven by CATSPER2 is required for fertilization. Proc. Natl Acad. Sci. USA 100, 14869–14874 (2003).
Qi, H. et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc. Natl Acad. Sci. USA 104, 1219–1223 (2007).
Jin, J. et al. Catsper3 and Catsper4 are essential for sperm hyperactivated motility and male fertility in the mouse. Biol. Reprod. 77, 37–44 (2007).
Nikpoor, P., Mowla, S. J., Movahedin, M., Ziaee, S. A. M. & Tiraihi, T. CatSper gene expression in postnatal development of mouse testis and in subfertile men with deficient sperm motility. Hum. Reprod. 19, 124–128 (2004).
Long, J. E., Lee, M. S. & Blithe, D. L. Male contraceptive development: update on novel hormonal and nonhormonal methods. Clin. Chem. 65, 153–160 (2019).
Cheng, C. Y. & Mruk, D. D. New frontiers in nonhormonal male contraception. Contraception 82, 476–482 (2010).
Kirichok, Y., Navarro, B. & Clapham, D. E. Whole-cell patch-clamp measurements of spermatozoa reveal an alkaline-activated Ca2+ channel. Nature 439, 737–740 (2006).
Tao, X., Lee, A., Limapichat, W., Dougherty, D. A. & MacKinnon, R. A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010).
Xie, J. F. et al. Structure of the human sodium leak channel NALCN in complex with FAM155A. Nat. Commun. 11, 5831 (2020).
Hagenbuch, B. & Stieger, B. The SLCO (former SLC21) superfamily of transporters. Mol. Aspects Med. 34, 396–412 (2013).
Fietz, D. et al. Membrane transporters for sulfated steroids in the human testis—cellular localization, expression pattern and functional analysis. PLoS One 8, e62638 (2013).
Suzuki, T. et al. Identification and characterization of novel rat and human gonad-specific organic anion transporters. Mol. Endocrinol. 17, 1203–1215 (2003).
Abbott, G. W. Chansporter complexes in cell signaling. FEBS Lett. 591, 2556–2576 (2017).
Li, N. N. et al. Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017).
Perez, G. I. et al. Prolongation of ovarian lifespan into advanced chronological age by Bax-deficiency. Nat. Genet. 21, 200–203 (1999).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. Model. 14, 354–360 (1996).
Yang, J. et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl Acad. Sci. USA 117, 1496–1503 (2020).
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D Biol. Crystallogr. 70, 2487–2499 (2014).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
DeLano, W. L. The PyMOL Molecular Graphics System http://www.pymol.org (2002).
Acknowledgements
We thank N. Yan and H. Yu for critical reading of the manuscript; M. Jiang and Y. Ru for help with the immunofluorescence experiments; the Cryo-EM Facility and HPC Center of Westlake University for providing cryo-EM and computation support; S. Feng and the Mass Spectrometry & Metabolomics Core Facility of Westlake University for protein sample MS analysis; and J. Bao, D. Wu and the Laboratory Animal Resources Center of Westlake University for help with animal maintenance and IVF experiments. This work was supported by Westlake Laboratory (Westlake Laboratory of Life Sciences and Biomedicine) (W101486022101) and an Institutional Startup Grant from the Westlake Education Foundation (101486021901) to J.W.
Author information
Authors and Affiliations
Contributions
J.W. conceived and supervised the project; S.L. and Y.Z. prepared the protein sample under the guidance of Z.Y. and J.W.; S.L. collected the cryo-EM data and M.K. calculated the cryo-EM map; J.W. built the model; all authors contributed to data analysis; and J.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Mouse genotyping, confocal imaging of spermatozoa and endogenous purification of the CatSpermasome from mouse sperm.
a, The genotype of each mouse was verified by two PCR reactions. Top, schematic of the genotyping procedure. Bottom, a group of representative PCR results. NC, negative control (empty template); WT, wild type; KI/+, heterozygote knock-in; KI/KI, homozygote knock-in. In total, 49 wild-type, 676 KI/+, and 207 KI/KI mice were verified. b, An drawing of a mouse sperm. CatSper was mainly distributed at the principal piece of spermatozoa. c, EGFP fluorescence was detected in the principal piece (red arrows) of knock-in mouse spermatozoa, but not wild-type mouse spermatozoa. Blue arrows, auto-fluorescence signal observed in the middle piece of spermatozoa. Shown here are one of the two images taken for each sample. Scale bars, 30 μm. d, Schematic of CatSpermasome purification. e, The purified protein sample was subjected to gel filtration analysis. The peak fractions of CatSpermasome (arrow) were collected and concentrated for cryo-EM and MS studies. Inset, the cross-linked protein sample was visualized on SDS–PAGE by silver staining. The corresponding protein band (arrow) in a separate gel without staining was cut out for MS analysis. For gel source data, see Supplementary Fig. 1. f, A representative EM micrograph of the CatSpermasome sample stained with uranyl acetate (one micrograph out of five in total for the negative staining sample). Scale bar, 50 nm.
Extended Data Fig. 2 Mass spectrometric analysis of the purified CatSpermasome.
a, MS samples were the same as used for the cryo-EM study. MS detected proteins are shown in order of decreasing confidence. All previously characterized CatSper components are listed (yellow). The top six entries with highest peptide spectrum match (PSM) values are all CATSPER proteins. Most of the contaminating proteins are cytoskeletal proteins. The newly identified components SLCO6C1 and CATSPERη are highlighted in red. TMEM249, which is probably another new component, is shaded in light blue. b, Representative MS spectra for specific peptides of CATSPER1, SLCO6C1, CATSPERη, and TMEM249.
Extended Data Fig. 3 Cryo-EM analysis of mouse CatSpermasome.
a, A representative motion-corrected micrograph of the CatSpermasome cryo-EM sample out of a dataset of 16,648 images. Scale bar, 50 nm. b, Two-dimensional class averages. Box size, 430 Å. c, Gold standard FSC curves for the 3D reconstructions. The curves were calculated with masks for the entire protein (overall map), and for masked regions of corresponding maps. See f for each map. d, Validation of the final structure models. FSC curves of the final refined model versus the summed map that it was refined against (black); of the model versus the first half-map (blue); and of the model versus the second half-map (red). The small difference between the blue and red curves indicates that the refinement of the atomic coordinates did not suffer from overfitting. e, Angular distribution of the particles of the final reconstruction generated by cryoSPARC. f, Flowchart of EM data processing (see ‘Image processing’ in Methods).
Extended Data Fig. 4 Structural features of the cytosolic regions.
a, The cytosolic region consists of two separate but interacting parts. Cytosolic map 1 is in close contact with the S6 segments of CATSPER2 and CATSPER3. A density surrounded in the bottom of cytosolic map 1 (shown in green) is connected to the density of the S6 segment of CATSPER3 and may belong to the carboxyl end of CATSPER3. Owing to limited resolution, the identity of cytosolic map 1 remains to be determined. Cytosolic map 2, however, is likely to be the subcomplex of EFCAB9 and CATSPERζ. The maps were generated in ChimeraX. b, Predicted structures of EFCAB9 by tFold and CATSPERζ by trRosseta. EFCAB9 has two EF-hand motif-containing lobes, which is very similar to calmodulin. CATSPERζ consists mainly of α-helices. c, Docking of the predicted structures of EFCAB9 and CATSPERζ into cytosolic map 2. The two lobes of EFCAB9 are in a compact conformation instead of the extended conformation in the predicted structure. The main body of CATSPERζ (light cyan in b) can be fitted into the remaining density near EFCAB9 in cytosolic map 2. Several fitted α-helices are indicated by arrows.
Extended Data Fig. 5 EM maps of CATSPER1–4.
a, Electron density maps of each segment of the transmembrane helices of CATSPER1–4. The boundaries of each displayed segment are labelled. The densities, shown as blue meshes, are contoured at 3–4σ in PyMOL. b, Electron density map of the selectivity filter and the pore helices. The densities are contoured at 4σ. Two tentatively assigned Na+ ions are shown as purple spheres. c, Electron density maps of detergent-like molecules. These densities may also belong to cholesterol or steroid hormones under physiological conditions. Three GDN molecules are tentatively assigned to these densities. The densities are found in a semi-open cavity formed by the S3, S4 and S4–5 segments of CATSPER1, but not CATSPER2–4, whose corresponding cavities are smaller. d, Structural superimposition of CATSPER1–4 indicates that CATSPER1 has a larger cavity for binding of detergent-like molecules.
Extended Data Fig. 6 Structural details and sequence alignment of the pore domain and VSDs.
a, Overall structure of the pore domain of CATSPER1–4. The critical DDDD residues in the selectivity filter are shown as sticks. Each S6 segment contains a π-helix turn (red arrows). b, Sequence alignment of the selectivity filter and the pore helices among mouse CATSPER1–4, human CATSPER1–4, and rabbit Cav1.1. The invariant Thr and Trp residues are shaded cyan. The DDDD residues in the selectivity filter of CATSPER1–4 and the corresponding residues EEEE in Cav1.1 are highlighted red. c, Sequence alignment of the VSDs among CATSPER1–4. The boundary of each segment is shaded light grey. Positively charged residues on S4 segments are shaded blue and residues corresponding to positions R1–R6 are boxed. An1 and CTC residues on segments S2 and S3 are shaded purple. d, Structural comparison of the VSDs among CATSPER1–4. The four VSDs are superimposed relative to CTC and An1 on S2. For visual clarity, the S1 segments are omitted and only the side chains of aligned residues and R4 residues on the S4 segments are shown.
Extended Data Fig. 7 Structure of the auxiliary subunits CATSPERβ, γ, δ and ε.
a, The overall structures of CATSPERβ, γ, δ and ε share similar domain organizations. The structures are shown in cartoon form and the sugar moieties in the glycosylation sites are shown as sticks. The NTDs, β-propeller domains, Ig-like domains, stem domains and transmembrane domains are coloured green, blue, orange, yellow and salmon, respectively. The head domain in CATSPERβ and the NTD2 domain in CATSPERε are coloured slate and cyan, respectively. b, Domain organization of CATSPERβ, γ, δ and ε. The boundaries for each domain and the identified glycosylation sites are labelled. The disulfide bonds are indicated by orange lines. See Extended Data Table 2 for details. c, Inter-subunit interactions among CATSPERβ, γ, δ and ε, shown in four side views. CATSPERβ is domain coloured and CATSPERγ, δ and ε are coloured as in Fig. 1. The side openings formed by two adjacent subunits are indicated by dotted lines. For visual clarity, the transmembrane helices are omitted. d, Extracellular view of the auxiliary subunits. The top of the channel is sealed by the Ig-like domains of CATSPERβ, γ, δ and ε. Bottom, close-up view of the interactions among the Ig-like domains (box in top image). The residues that mediate the interactions in the interface are shown as sticks. Hydrogen bonds are indicated by red dashed lines. e, Interactions between the transmembrane domains of CATSPERβ, γ, δ and ε and the adjacent VSDs of CATSPER1–4. The residues that contribute to the interface interactions are shown as sticks. Potential hydrogen bonds are indicated by red dashed lines. The stem domains of CATSPERβ and CATSPERε are further from the adjacent channel subunits than those of CATSPERγ and CATSPERδ (double-headed arrows).
Extended Data Fig. 8 Representative EM maps of the auxiliary subunits CATSPERβ, γ, δ and ε.
The selected segments cover almost every domain of CATSPERβ, γ, δ and ε. Representative densities for the glycosylation sites from each subunit are also presented. The densities, shown as blue meshes, are contoured at 4–5σ in PyMOL.
Extended Data Fig. 9 Structural analysis and sequence alignment of SLCO6C1.
a, Side view (left) and central slice view (right) of the electron density map of SLCO6C1 and the adjacent components. The map was generated in ChimeraX. b, SLCO6C1 is captured in an outward-facing conformation. The structure is shown in cylindrical helices cartoon mode and coloured by domain. Right, the N and C domains of SLCO6C1 each contain six transmembrane helices and are pseudo-symmetric, with an r.m.s.d. of ~6 Å when superimposed. c, Sequence alignment among mouse SLCO6C1, rat SLCO6C1 and human SLCO6A1. The glycosylation site (red triangle) and the residues that may be involved in interactions with CATSPERε (yellow triangles) are conserved among species. The uniport IDs for the aligned sequences are: mSLCO6C1: Q3V161; rSLCO6C1: G3V7R7; and hSLCO6A1: Q86UG4.
Extended Data Fig. 10 Structural characterization of CATSPERη and TMEM249.
a, Side view (left) and central slice view (right) of the electron density maps of CATSPERη and the adjacent components. The map was generated in ChimeraX. b, Electron density maps of CATSPERη. The map allows accurate assignment of the side chains of the bulky residues. The densities, shown as blue meshes, are contoured at 4σ in PyMOL. c, Predicted structure of TMEM249 by tFold perfectly fits into the density near CATSPERη with minor adjustments. The structure has the characteristics of two transmembrane helices and a β-sheet (arrows) in the cytosolic domain. d, TMEM249 interacts with CATSPERη and VSD4 in the transmembrane and cytosolic regions, respectively. Unlike CATSPERη, TMEM249 does not interact with the stem domain of CATSPERβ.
Extended Data Fig. 11 Immunofluorescence detection of representative CatSpermasome components in wild-type sperm.
CATSPER1, CATSPER4, CATSPERβ and TMEM249 are mainly distributed in the principal piece of sperm (red fluorescence signal). The sperm head, middle piece and principal piece are indicated by black, blue, and red arrows, respectively. NC, negative control (without primary antibody). Shown here are one of three images taken for each sample. Scale bars, 20 μm.
Supplementary information
Supplementary Figure 1
Uncropped raw data gel from Extended Data Fig. 1 - SDS-PAGE gel for Extended Data Fig. 1e showing the BS3-crosslinked purified mouse CatSpermasome sample by silver staining.
Video 1
: Overall EM map of the mouse CatSpermasome The displayed overall composite map was in the same color scheme as Fig. 1. The movie was generated in ChimeraX
Rights and permissions
About this article
Cite this article
Lin, S., Ke, M., Zhang, Y. et al. Structure of a mammalian sperm cation channel complex. Nature 595, 746–750 (2021). https://doi.org/10.1038/s41586-021-03742-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03742-6
This article is cited by
-
Control of intracellular pH and bicarbonate by CO2 diffusion into human sperm
Nature Communications (2023)
-
Cryo-EM structures of human organic anion transporting polypeptide OATP1B1
Cell Research (2023)
-
Advances in the study of genetic factors and clinical interventions for fertilization failure
Journal of Assisted Reproduction and Genetics (2023)
-
A novel Human Conception Optimizer for solving optimization problems
Scientific Reports (2022)
-
3D structure and in situ arrangements of CatSper channel in the sperm flagellum
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.