Abstract
Hepatocellular carcinoma (HCC)—the most common form of liver cancer—is an aggressive malignancy with few effective treatment options1. Lenvatinib is a small-molecule inhibitor of multiple receptor tyrosine kinases that is used for the treatment of patients with advanced HCC, but this drug has only limited clinical benefit2. Here, using a kinome-centred CRISPR–Cas9 genetic screen, we show that inhibition of epidermal growth factor receptor (EGFR) is synthetic lethal with lenvatinib in liver cancer. The combination of the EGFR inhibitor gefitinib and lenvatinib displays potent anti-proliferative effects in vitro in liver cancer cell lines that express EGFR and in vivo in xenografted liver cancer cell lines, immunocompetent mouse models and patient-derived HCC tumours in mice. Mechanistically, inhibition of fibroblast growth factor receptor (FGFR) by lenvatinib treatment leads to feedback activation of the EGFR–PAK2–ERK5 signalling axis, which is blocked by EGFR inhibition. Treatment of 12 patients with advanced HCC who were unresponsive to lenvatinib treatment with the combination of lenvatinib plus gefitinib (trial identifier NCT04642547) resulted in meaningful clinical responses. The combination therapy identified here may represent a promising strategy for the approximately 50% of patients with advanced HCC who have high levels of EGFR.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request. The RNA-sequencing data have been deposited in the Gene Expression Omnibus (GSE157905). Source data are provided with this paper.
References
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
Kudo, M. et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173 (2018).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Zucman-Rossi, J., Villanueva, A., Nault, J.-C. & Llovet, J. M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239 (2015).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008).
Finn, R. S. et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382, 1894–1905 (2020).
Evers, B. et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 34, 631–633 (2016).
Zhu, A. X. et al. SEARCH: a phase III, randomized, double-blind, placebo-controlled trial of sorafenib plus erlotinib in patients with advanced hepatocellular carcinoma. J. Clin. Oncol. 33, 559–566 (2015).
Matsuki, M. et al. Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models. Cancer Med. 7, 2641–2653 (2018).
Wagle, M.-C. et al. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis. Oncol. 2, 7 (2018).
Pirker, R. et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 13, 33–42 (2012).
Kimura, T. et al. Immunomodulatory activity of lenvatinib contributes to antitumor activity in the Hepa1-6 hepatocellular carcinoma model. Cancer Sci. 109, 3993–4002 (2018).
Li, E., Lin, L., Chen, C. W. & Ou, D. L. Mouse models for immunotherapy in hepatocellular carcinoma. Cancers (Basel) 11, 1800 (2019).
Keng, V. W. et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 27, 264–274 (2009).
He, H. et al. Platelet-derived growth factor requires epidermal growth factor receptor to activate p21-activated kinase family kinases. J. Biol. Chem. 276, 26741–26744 (2001).
Lee, S.-W. et al. EGFR–Pak signaling selectively regulates glutamine deprivation-induced macropinocytosis. Dev. Cell 50, 381–392 (2019).
Vaseva, A. V. et al. KRAS suppression-induced degradation of MYC is antagonized by a MEK5–ERK5 compensatory mechanism. Cancer Cell 34, 807–822 (2018).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Lito, P. et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012).
Kosoff, R. E. et al. Pak2 restrains endomitosis during megakaryopoiesis and alters cytoskeleton organization. Blood 125, 2995–3005 (2015).
Kobayashi, S. et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res. 66, 11389–11398 (2006).
Philip, P. A. et al. Phase II study of erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J. Clin. Oncol. 23, 6657–6663 (2005).
Chan, S. L. et al. New utility of an old marker: serial α-fetoprotein measurement in predicting radiologic response and survival of patients with hepatocellular carcinoma undergoing systemic chemotherapy. J. Clin. Oncol. 27, 446–452 (2009).
Llovet, J. M. & Lencioni, R. mRECIST for HCC: performance and novel refinements. J. Hepatol. 72, 288–306 (2020).
Ding, X. et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology 157, 1630–1645 (2019).
Cremolini, C. et al. Early tumor shrinkage and depth of response predict long-term outcome in metastatic colorectal cancer patients treated with first-line chemotherapy plus bevacizumab: results from phase III TRIBE trial by the Gruppo Oncologico del Nord Ovest. Ann. Oncol. 26, 1188–1194 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Jin, H. et al. Regulator of calcineurin 1 gene isoform 4, down-regulated in hepatocellular carcinoma, prevents proliferation, migration, and invasive activity of cancer cells and metastasis of orthotopic tumors by inhibiting nuclear translocation of NFAT1. Gastroenterology 153, 799–811 (2017).
Mazières, J. et al. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: re-analysis of the SATURN study. Lung Cancer 82, 231–237 (2013).
Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014).
Wang, L. et al. High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep. 21, 773–783 (2017).
Schuhmacher, M. et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29, 397–406 (2001).
Wang, C. et al. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature 574, 268–272 (2019).
World Medical Association. World Medical Association Declaration of Helsinki. Ethical principles for medical research involving human subjects. Bull. World Health Organ. 79, 373–374 (2001).
Acknowledgements
We thank R. G. H. Beets-Tan for discussion and assistance in MRI image analysis. This work was funded by grants from the European Research Council (ERC 787925 to R.B.), the Dutch Cancer Society through the Oncode Institute, the National Natural Science Foundation of China (82073039 to H.J., 82011530441 to H.J., 81702838 to H.J., 81920108025 to W.Q. and 81801800 to Y.S.), National Science and Technology Key Project of China (2018ZX10732202-002-003 to W.Q. and 2018ZX10302205-002-003 to H.J.), Shanghai Rising-Star Program (19QA1408200 to H.J.), Shanghai Municipal Science and Technology Project (20JC1411100 to W.Q.), 111 project (B21024 to W.Q.), Shanghai Municipal Commission of Health and Family Planning (2018YQ20 to H.J.) and Shanghai Jiao Tong University School of Medicine (YG2019GD01 to H.J. and PY11-17-013 to H.J.). We thank the facilities of The Netherlands Cancer Institute.
Author information
Authors and Affiliations
Contributions
R.B., W.Q., B.Z. and W.Z. supervised all of the research. R.B., W.Q. and H.J. wrote the manuscript. H.J., Y.L., S.Y., C.F.A.R., S.W., F.J., A.B., Y.Z., J.L., H. Wang, Y.G., X.Z., M.H.P.d.G., S.V. and N.I. designed and performed the preclinical experiments. H.J., C.L. and R.L.B. performed data analysis. B.Z., Y.S. and D.C. designed and performed the clinical study. L.W., C.W., M.H.D., Y.Y., E.M.H., H. Wu, C.S., R.L.B. and L.A. provided advice and technical support for the project. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
H.J. and R.B. are listed as inventors on a patent application using the drug combination described here. The other authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 A kinome CRISPR screen in the lenvatinib-resistant liver cancer cell line SNU449.
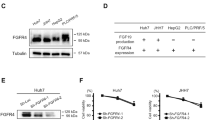
a, Liver cancer cells are resistant to lenvatinib treatment in vitro. Short-term viability assay of liver cancer cell lines. Cells were treated with increasing concentrations of lenvatinib for 72 h, and cell viability was determined using CellTiter-Blue. Data are mean ± s.e.m. Cell lines with half-maximum inhibitory concentration (IC50) values < 5 μM are classed as relatively sensitive cells, whereas cell lines with IC50 > 5 μM are classed as relatively resistant cells. SNU449, JHH1, SNU182, PLC/PRF5, MHCC97H, HepG2, SNU398, Huh7 and Hep3B, n = 8; Huh6 and SK-Hep1, n = 7. b, Scatter plots of log10-transformed normalized read counts of independent biological replicates of GeCKO library lentiviral transductions with R2 values shown (n = 3). Replicates from triplicate transductions showed good correlation at the sgRNA level. T0, SNU449 cells after puromycin selection; Tuntreated, SNU449 cells cultured for 14 days without drug treatment; Ttreated, SNU449 cells cultured for 14 days with 10 μM lenvatinib. c, Fold change distribution of all gRNAs targeting essential genes or nonessential genes in three independent biological replicates. The sgRNAs for essential genes were strongly depleted relative to the sgRNAs targeting nonessential genes, highlighting the quality of the screen. ‘p’, 50 sgRNAs targeting 10 essential genes (red); ‘n’, 50 non-targeting control sgRNAs (blue); ‘x’, 5,860 sgRNAs targeting 504 human kinases (grey). d, Average log2-transformed fold change in each individual EGFR gRNA in the pooled CRISPR library (n = 3 independent experiments). Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test
Extended Data Fig. 2 Synthetic lethal effects of lenvatinib and EGFR inhibitors on liver cancer cells expressing a high level of EGFR in vitro.
a–f, EGFRhigh liver cancer cell lines (SNU449, JHH1, Huh6 and SNU182) or EGFRlow cells (SNU398 and HepG2) were treated with lenvatinib, EGFR inhibitors (gefitinib or erlotinib) or their combination at the indicated concentrations. Growth curves were determined by Incucyte live cell analyses. Data are mean ± s.e.m., n = 5. g–l, The quantification of three independent assays in Fig. 2b–g. Data are mean ± s.e.m., n = 3 independent experiments. m–o, Ectopic EGFR expression confers resistance to lenvatinib in EGFRlow cells. m, Western blot analysis of p-EGFR and total EGFR levels in SNU398 and HepG2 cells infected with control or EGFR overexpression vector. HSP90 served as a control. n, o, SNU398 and HepG2 cells expressing control vector or EGFR were cultured with or without lenvatinib (SNU398, 5 μM; HepG2, 10 μM). n, The cells were fixed, stained and photographed after 14 days. o, Quantification of three independent assays. Data are mean ± s.e.m., n = 3 independent experiments
Extended Data Fig. 3 The combination of sorafenib and the EGFR inhibitor gefitinib shows no synergy in liver cancer cells in vitro.
a–f, Liver cancer cell lines were treated with sorafenib, the EGFR inhibitor gefitinib or their combination at the indicated concentrations. The cells were fixed and stained after 10–14 days. Representative data from three independent experiments. g–l, The quantification of the three independent assays shown in a–f. Data are mean ± s.e.m., n = 3 independent experiments
Extended Data Fig. 4 Bliss independence model evaluating the synergistic effect between lenvatinib or sorafenib with gefitinib in liver cancer cells.
The Bliss independence model was applied to the quantification data of colony-formation experiments in Extended Data Figs. 2g–l, 3g–l. The data in the untreated condition (no lenvatinib or gefitinib) were normalized as 0, indicating no inhibition of cell viability. Then, the additive scores (blissAdd) were generated by multiplying the normalized effect of the two single drugs. We further calculated the reduction in viability by subtracting the mean of the blissAdd from the mean of the measured combination scores (measured). The green background indicates the synergistic effect with P ≤ 0.05 and when the reduction in viability is more than 10%. a–f, Bliss independence analysis for the combination of gefitinib and lenvatinib shown in Extended Data Fig. 2g–l. g–l, Bliss independence analysis for the combination of gefitinib and sorafenib shown in Extended Data Fig. 3g–l. Data are mean ± s.e.m., n = 3 independent experiments. P values were determined by unpaired two-sided Student’s t-test
Extended Data Fig. 5 EGFR inhibitors increased the sensitivity of EGFRhigh liver cancer cells to FGFR inhibitors in vitro.
a, b, Long-term colony formation assay of 11 liver cancer cell lines treated with the FGFR inhibitors AZD4547 (a) and BGJ398 (b). Cells were grown in the absence or presence of drugs at the indicated concentrations for 10–14 days, fixed and stained. c–f, EGFRhigh liver cancer cell lines (SNU449, JHH1, Huh6 and SNU182) were treated with FGFR inhibitors (AZD4547 or BGJ398), EGFR inhibitor (gefitinib) or their combination at the indicated concentrations. Growth curves were determined by Incucyte live cell analyses. Data are mean ± s.e.m., n = 5. g–l, FGFR inhibitors (AZD4547 or BGJ398) but not the FGFR4-specific inhibitor BLU-554 showed a synergistic effect with EGFR inhibitors in EGFRhigh liver cancer cells in vitro. Liver cancer cell lines with different EGFR expression levels were treated with FGFR inhibitors (AZD4547 or BGJ398), FGFR4-specific inhibitor (BLU-554), EGFR inhibitors (gefitinib or erlotinib) or their different combinations at the indicated concentrations. The cells were fixed and stained after 10–14 days. m–o, SNU449 and JHH1 cells were treated with gefitinib (2.5 μM for SNU449; 0.625 μM for JHH1), BGJ398, sorafenib, or as a combination of two or three drugs. m, The cells were fixed, stained and photographed after 14 days. n, o, Quantification of three independent assays. Data are mean ± s.e.m., n = 3 independent experiments. P values were determined by two-sided unpaired Student’s t-test
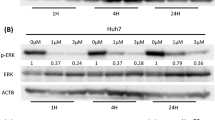
Extended Data Fig. 6 Lenvatinib plus EGFR inhibitors suppresses the ERK1/2 MAPK pathway in EGFRhigh liver cancer cells.
a–f, Immunoblot and quantification of p-EGFR and p-ERK1/2 levels in liver cancer cells after the indicated treatment. EGFRhigh and EGFRlow liver cancer cell lines were treated with lenvatinib, EGFR inhibitors (gefitinib or erlotinib) or their combination at the indicated concentrations for 24 h. Protein extracts were probed with specific antibodies against EGFR (total and phosphorylated), ERK1/2 (total and phosphorylated) and HSP90 (as a loading control). Data are mean ± s.e.m., n = 3 independent experiments. P values were determined by two-sided unpaired Student’s t-test. g–j, Liver cancer cell lines SNU449 (g), JHH1 (h), Huh6 (i) and SNU182 (j) were treated with DMSO, lenvatinib, gefitinib or the combination of both drugs for 24 h. The mRNA levels of 10 transcriptional targets of MAPK signalling (DUSP4, DUSP6, ETV4, ETV5, EPHA2, EPHA4, SPRY2, SPRY4, PHLDA1 and CCND1) were determined by RT–qPCR analysis. ACTB (which encodes β-actin) was used as internal control. Data are mean ± s.e.m. (n = 3)
Extended Data Fig. 7 The combination of lenvatinib and gefitinib suppresses tumour growth without toxicity in cell-line-derived immunodeficient mouse models in vivo.
a–j, Liver cancer cells were grown as tumour xenografts in BALB/c nude mice. After tumour establishment (~200 mm3), mice were treated with vehicle, lenvatinib (4 mg kg−1), gefitinib (80 mg kg−1) or lenvatinib (4 mg kg−1) plus gefitinib (80 mg kg−1), for the indicated number of days. Representative images of H&E, PCNA, Ki-67, cleaved caspase-3 and CD31 staining in SNU449 (a) and Huh6 (b) xenograft models. Scale bars, 50 μm. c–j, Quantification of PCNA+ cells (c, g), Ki-67+ cells (d, h), cleaved caspase-3+ cells (e, i) and microvessel density (MVD) (f, j) per high-power field in representative sections from each group. n = 6 per group. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. k, l, The body weight of mice in the SNU449 (k) and Huh6 (l) xenograft models. Data are mean ± s.e.m. n = 6 mice per group
Extended Data Fig. 8 The combination of lenvatinib and gefitinib suppresses tumour growth and extends survival time in HCC PDX mouse models in vivo.
a, b, Four HCC PDX models were established and examined for EGFR expression by IHC analyses. a, Representative images of H&E and EGFR staining in the four PDX models. Scale bars, 50 μm. b, H-score of EGFR expression levels in the four PDX models. Data are mean ± s.e.m., n = 5 samples. c–n, After tumour establishment (~200 mm3), mice were treated with vehicle, lenvatinib (4 mg kg−1), gefitinib (80 mg kg−1) or lenvatinib (4 mg kg−1) plus gefitinib (80 mg kg−1). c–f, Growth curves of four HCC PDX models in mice treated with vehicle, lenvatinib, gefitinib or their combination. n = 5 mice per group. Data are mean ± s.e.m. P values were determined by two-way ANOVA with Tukey multiple comparisons. g–j, Kaplan–Meier survival curves of the PDX models after the indicated treatments. n = 5 mice per group. P values were determined by log-rank Mantel–Cox test. k–n, Body weights of mice in the PDX models with the aforementioned treatments were assessed. n = 5 mice per group. Data are mean ± s.e.m
Extended Data Fig. 9 The combination of lenvatinib and gefitinib suppresses tumour growth without toxicity in immunocompetent mouse models in vivo.
a, Western blot analysis of total EGFR levels in mouse liver tissues and mouse liver cancer cells (Hepa1-6). Representative of three independent experiments. b, Hepa1-6 cells were treated with lenvatinib, the EGFR inhibitor gefitinib or their combination at the indicated concentrations. The cells were fixed and stained after 7 days. Representative of three independent experiments. c, After intrahepatic inoculation of Hepa1-6 cells in C57BL/6 mice, mice were treated with vehicle, lenvatinib (4 mg kg−1), gefitinib (80 mg kg−1) or lenvatinib (4 mg kg−1) plus gefitinib (80 mg kg−1) for 2 weeks. Tumour weight was measured. Data are mean ± s.e.m. n = 6 mice per group. P values were determined by two-sided unpaired Student’s t-test. d, Body weights of mice in the Hepa1-6 orthotopic models with the aforementioned treatments were assessed. n = 6 mice per group. Data are mean ± s.e.m. e, Schematic of gene delivery by hydrodynamic tail vein injection (HDTVi) of the Myc proto-oncogene transposon system and a CRISPR–Cas9 vector targeting the Trp53 tumour suppressor, which was used to induce HCC 2–3 weeks after hydrodynamic tail vein injections. f, Western blot analysis of total EGFR levels in mouse liver tissues and mouse liver cancer cells (MycOETrp53KO). Representative of three independent experiments. g, MycOETrp53KO mouse liver cancer cells were treated with lenvatinib, EGFR inhibitor gefitinib or their combination at the indicated concentrations, respectively. The cells were fixed and stained after 5 days. Representative of three independent experiments. h, Survival curve generated from mice bearing MycOETrp53KO tumours, treated with vehicle (n = 6; median survival of 11 days), lenvatinib (4 mg kg−1; n = 9; median survival of 23 days), gefitinib (80 mg kg−1; n = 6; median survival of 13.5 days) or lenvatinib plus gefitinib (n = 9; median survival of 31 days). P values were determined by two-sided log-rank test. i, Body weights of mice in the MycOETrp53KO mouse liver cancer models with the aforementioned treatments were assessed. n = 6–9 mice per group. Data are mean ± s.e.m. j, k, Mice bearing MycOETrp53KO tumours treated with vehicle, lenvatinib (4 mg kg−1), gefitinib (80 mg kg−1) or a combination of both drugs were killed at the end point after treatment. Tumours were dissociated as single-cell suspensions, and flow cytometry analyses were performed to determine the content of tumour-associated lymphoid cells (NK cells, CD8+ T cells, CD4+ T cells and regulatory T cells (Tregs)) and myeloid cells (monocytes, neutrophils, tumour-associated macrophages (TAMs) and dendritic cells (DCs)). Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. Sample sizes are given in the Methods
Extended Data Fig. 10 Concurrent inhibition of ERK5 and ERK1/2 shows an enhanced anti-tumour effect in vitro.
a, Quantification of Fig. 3b with three independent experiments. The phosphorylation levels of each protein were normalized based on their total protein levels. n = 3 independent experiments. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. b, Western blot analysis of EGFR–PAK2–ERK5 cascade in JHH1 cells, treated with lenvatinib, EGFR inhibitors (gefitinib and erlotinib) or their combination at the indicated concentrations for 6 h. HSP90 served as a loading control. c, The phosphorylation levels of each protein in b were normalized based on their total protein levels. n = 3 independent experiments. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. d, EGFR expression of SNU449 cells was knocked down by two independent shRNAs, and cells were further treated with lenvatinib (5 μM) for 6 h. Western blot analysis of EGFR–PAK2–ERK5 cascade was performed. HSP90 served as a loading control. The pLKO vector was used in the control experiment. e, Quantification of Fig. 3c with three independent experiments. The phosphorylation levels of each protein were normalized based on their total protein levels. n = 3 independent experiments. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. f, JHH1 cells were treated with lenvatinib (5 μM), PAK inhibitor FRAX1036 (2.5 μM) or their combination for 6 h, and western blot analysis was performed with the indicated antibodies. g, The phosphorylation levels of each protein in f were normalized based on their total protein levels. n = 3 independent experiments. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test. h, i, Long-term colony formation assays showing synergistic effects of lenvatinib and the PAK inhibitor FRAX1036 on the proliferation of SNU449 (h) and JHH1 (i) cells. Representative of three independent experiments. j, k, Long-term colony formation assays showing synergistic effects of lenvatinib and ERK5 inhibitor XMD8-92 on the proliferation of SNU449 (j) and JHH1 (k) cells. Representative of three independent experiments. l, m, SNU449 (l) and JHH1 (m) cells were treated with lenvatinib (5 μM), the ERK5 inhibitor XMD8-92 (2.5 μM) or their combination for 6 h, and western blot analysis was performed with the indicated antibodies. HSP90 served as a loading control. n–q, Knockout of ERK5 using the CRISPR–Cas9 system enhances the response to lenvatinib. The ERK5 knockout efficiency was determined by western blot in SNU449 (n) and JHH1 (p) cells. HSP90 served as a loading control. The effects of ERK5 knockout on proliferation were indicated by colony formation. The ERK5 knockout SNU449 (o) and JHH1 (q) cells, or their respective control cells were treated with DMSO or 5 μM lenvatinib. After 10 days of culture, cells were fixed, stained and photographed. NT, non-targeting sgRNA. r, Quantification of IHC staining in Fig. 3d. n = 6 per group. Data are mean ± s.e.m. P values were determined by two-sided unpaired Student’s t-test
Extended Data Fig. 11 RNA-sequencing analysis of the combination treatment of lenvatinib and gefitinib in EGFRhigh liver cancer cells.
a–d, Heat-map representations of the log2-transformed gene expression changes (log2FC) in liver cancer cells SNU449 (a), JHH1 (b), Huh6 (c) and SNU182 (d) treated with DMSO, 2.5 μM gefitinib, 5 μM lenvatinib, or the two drugs for 24 h. Differential expression of the whole genome is shown on the basis of each treatment versus DMSO using RNA-sequencing analysis. To show robust changes, only genes that have a mean read count over all samples equal or greater than 100 were included. To prevent too much dilution of the colour-coding, the log2-transformed fold-change values were truncated in that lower than −2 were set to −2 and higher than 2 were set to 2. Heat maps depicting all three treatments were then generated by unsupervised hierarchical clustering. One replicate was used for each sample. e, For the genes with a mean read count over all samples greater than 100 per line, the top 25% highest and 25% lowest synergy scores were determined and shown as heat maps. These lists were merged and an unsupervised hierarchical clustering was performed. f, GSEA analysis of the gene set of Kobayashi ‘EGFR signalling down’ in each comparison. Unsupervised hierarchical clustering of normalized enrichment scores (NES) was used to generate a comprehensive heat-map visualization of the functional transcriptional outputs of the four cell lines. *P < 0.001. g, GSEA analysis of curated gene sets was performed, Kobayashi ‘EGFR signalling down’ and Schuhmacher ‘MYC targets up’ were identified as two of the highest-ranking downregulated gene sets in the combination-treated cells in all four liver cancer cell lines based on additional gene alterations in combination group. h–k, A fold change preranked list of each treatment versus DMSO was used to run GSEA against the Hallmark gene sets in SNU449 (h), JHH1 (i), Huh6 (j) and SNU182 (k) cells. Unsupervised hierarchical clustering of normalized enrichment scores was used to generate a comprehensive heat-map visualization of the functional transcriptional outputs of each treatment (FDR < 0.1). l–n, GSEA indicates that the gene sets of Hallmark ‘MYC targets v1’ (l), Hallmark ‘MYC targets v2’ (m) and Hallmark ‘KRAS signalling up’ (n) were negatively enriched in the combination group based on additional gene alterations.
Extended Data Fig. 12 Expression and prognostic effect of EGFR in patients with HCC.
a–d, IHC staining analyses of EGFR were performed in 298 patients with HCC. The H-score method assigned a score of 0–300 to each patient sample, based on the percentage of cells stained at different intensities. The discriminatory threshold was set at 200. All samples were classified as low (H < 200; EGFRlow) or high (H ≥ 200; EGFRhigh) EGFR expression. a, Typical images of EGFRlow and EGFRhigh cases according to their H-score. Scale bars, 200 μm. b, Distribution of EGFR expression levels in 298 patients with HCC as represented by various H-score ranges. Out of 298 patients with HCC, 157 had H-score of H ≥ 200 (EGFRhigh). Kaplan–Meier analysis for overall survival (OS, c) and time to relapse (TTR, d) was performed according to EGFR levels. Statistical analysis by log-rank Mantel–Cox test. e, The CONSORT diagram shows the patient flow through the clinical trial. f, H&E and IHC analyses showing high EGFR expression in formalin-fixed paraffin embedded (patient IDs A–L) tissue sections from HCC. All of the biopsies were obtained from liver tumours when patients were diagnosed or treated with primary hepatectomy. Scale bars, 50 μm
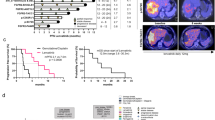
Extended Data Fig. 13 Clinical response of patients with lenvatinib-resistant advanced HCC to the combination of lenvatinib plus gefitinib.
a, Schematic representation of the treatment timeline of patient A with HCC after diagnosis. The dates and treatments administered are annotated along the arrow. MRI scans of patient A were performed during lenvatinib monotherapy (IM-T1), after lenvatinib monotherapy (IM-T2) and after combination therapy (IM-T3). The red label indicates the duration of lenvatinib treatment; the green label indicates the duration of gefitinib treatment. b, Serum AFP level of patient A was monitored during lenvatinib monotherapy (AFP-T1), after lenvatinib monotherapy (AFP-T2), and after combination therapy of lenvatinib plus gefitinib (AFP-T3). c, Schematic representation of the treatment timeline of patient C with HCC after diagnosis. d, Serum AFP level of patient C during the indicated treatment. e–n, Serum AFP level of patients B, D–L was monitored before or during lenvatinib monotherapy (AFP-T1), after lenvatinib monotherapy (AFP-T2) and after combination therapy of lenvatinib plus gefitinib (AFP-T3). o–x, MRI scans of patients with HCC were performed before or during lenvatinib monotherapy (IM-T1), after lenvatinib monotherapy (IM-T2) and after combination therapy of lenvatinib plus gefitinib (IM-T3). The tumour size of the major target lesions measured by MRI scan in patients with HCC was plotted on the y axes against indicated time point and treatment duration on the x axes
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-2 (the uncropped blots and FACS gating strategy) and the trial protocol.
Supplementary Table 1
Characteristics of 12 recruited HCC patients.
Supplementary Table 2
Kinome-centred CRISPR screen in SNU449 cells.
Supplementary Table 3
Primer sequences used in qRT-PCR analyses.
Supplementary Table 4
Information for antibodies used in the Flow cytometry assays.
Source data
Rights and permissions
About this article
Cite this article
Jin, H., Shi, Y., Lv, Y. et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature 595, 730–734 (2021). https://doi.org/10.1038/s41586-021-03741-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03741-7
This article is cited by
-
Identification of a DNA damage repair-related LncRNA signature for predicting the prognosis and immunotherapy response of hepatocellular carcinoma
BMC Genomics (2024)
-
RARRES1 inhibits hepatocellular carcinoma progression and increases its sensitivity to lenvatinib through interaction with SPINK2
Biology Direct (2024)
-
Ferroptosis in cancer: From molecular mechanisms to therapeutic strategies
Signal Transduction and Targeted Therapy (2024)
-
RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives
Nature Reviews Gastroenterology & Hepatology (2024)
-
Deep learning enables the discovery of a novel cuproptosis-inducing molecule for the inhibition of hepatocellular carcinoma
Acta Pharmacologica Sinica (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.