Abstract
Communication within the glial cell ecosystem is essential for neuronal and brain health1,2,3. The influence of glial cells on the accumulation and clearance of β-amyloid (Aβ) and neurofibrillary tau in the brains of individuals with Alzheimer’s disease (AD) is poorly understood, despite growing awareness that these are therapeutically important interactions4,5. Here we show, in humans and mice, that astrocyte-sourced interleukin-3 (IL-3) programs microglia to ameliorate the pathology of AD. Upon recognition of Aβ deposits, microglia increase their expression of IL-3Rα—the specific receptor for IL-3 (also known as CD123)—making them responsive to IL-3. Astrocytes constitutively produce IL-3, which elicits transcriptional, morphological, and functional programming of microglia to endow them with an acute immune response program, enhanced motility, and the capacity to cluster and clear aggregates of Aβ and tau. These changes restrict AD pathology and cognitive decline. Our findings identify IL-3 as a key mediator of astrocyte–microglia cross-talk and a node for therapeutic intervention in AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA-seq data for microglia from Il3–/–5xFAD and 5xFAD mice have been deposited to NCBI-GEO under accession GSE163289. RNA-seq data for WT, 5xFAD, Trem2−/− and Trem2−/−5xFAD mice have been previously described24 and were deposited to GEO: GSE132508. scRNA sequencing data for homeostatic, DAM stage 1, and DAM stage 2 microglia have been previously described23 and were deposited to GEO: GSE98969. Source data are provided with this paper.
References
Linnerbauer, M., Wheeler, M. A. & Quintana, F. J. Astrocyte crosstalk in CNS inflammation. Neuron 108, 608–622 (2020).
Vainchtein, I. D. & Molofsky, A. V. Astrocytes and microglia: in sickness and in health. Trends Neurosci. 43, 144–154 (2020).
Castellani, G. & Schwartz, M. Immunological features of non-neuronal brain cells: implications for Alzheimer’s disease immunotherapy. Trends Immunol. 41, 794–804 (2020).
Fakhoury, M. Microglia and astrocytes in Alzheimer’s disease: implications for therapy. Curr. Neuropharmacol. 16, 508–518 (2018).
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
Mindur, J. E. & Swirski, F. K. Growth factors as immunotherapeutic targets in cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 39, 1275–1287 (2019).
Gómez Ravetti, M. & Moscato, P. Identification of a 5-protein biomarker molecular signature for predicting Alzheimer’s disease. PLoS ONE 3, e3111 (2008).
Ray, S. et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat. Med. 13, 1359–1362 (2007).
Britschgi, M. et al. Modeling of pathological traits in Alzheimer’s disease based on systemic extracellular signaling proteome. Mol. Cell. Proteomics 10, 008862 (2011).
Soares, H. D. et al. Plasma biomarkers associated with the apolipoprotein E genotype and Alzheimer disease. Arch. Neurol. 69, 1310–1317 (2012).
Huberman, M. et al. Correlation of cytokine secretion by mononuclear cells of Alzheimer patients and their disease stage. J. Neuroimmunol. 52, 147–152 (1994).
Kiddle, S. J. et al. Plasma based markers of [11C] PiB-PET brain amyloid burden. PLoS ONE 7, e44260 (2012).
Frei, K., Bodmer, S., Schwerdel, C. & Fontana, A. Astrocytes of the brain synthesize interleukin 3-like factors. J. Immunol. 135, 4044–4047 (1985).
Frei, K., Bodmer, S., Schwerdel, C. & Fontana, A. Astrocyte-derived interleukin 3 as a growth factor for microglia cells and peritoneal macrophages. J. Immunol. 137, 3521–3527 (1986).
Zambrano, A., Otth, C., Maccioni, R. B. & Concha, I. I. IL-3 controls tau modifications and protects cortical neurons from neurodegeneration. Curr. Alzheimer Res. 7, 615–624 (2010).
Zambrano, A., Otth, C., Mujica, L., Concha, I. I. & Maccioni, R. B. Interleukin-3 prevents neuronal death induced by amyloid peptide. BMC Neurosci. 8, 82 (2007).
Herisson, F. et al. Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci. 21, 1209–1217 (2018).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Zenaro, E. et al. Neutrophils promote Alzheimer’s disease-like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 21, 880–886 (2015).
Pasciuto, E. et al. Microglia require CD4 T cells to complete the fetal-to-adult transition. Cell 182, 625–640 (2020).
Anzai, A. et al. Self-reactive CD4+ IL-3+ T cells amplify autoimmune inflammation in myocarditis by inciting monocyte chemotaxis. J. Exp. Med. 216, 369–383 (2019).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Griciuc, A. et al. TREM2 acts downstream of CD33 in modulating microglial pathology in Alzheimer’s disease. Neuron 103, 820–835 (2019).
Choi, S. H. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
Park, J. et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 21, 941–951 (2018).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Weber, G. F. et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 347, 1260–1265 (2015).
Turnbull, I. R. et al. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177, 3520–3524 (2006).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Hsiau, T. et al. Inference of CRISPR edits from Sanger trace data. Preprint at https://doi.org/10.1101/251082 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012).
Robbins, C. S. et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172 (2013).
DeVos, S. L. & Miller, T. M. Direct intraventricular delivery of drugs to the rodent central nervous system. J. Vis. Exp. 75, e50326 (2013).
McQuade, A. et al. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13, 67 (2018).
Griciuc, A. et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78, 631–643 (2013).
Kraeuter, A. K., Guest, P. C. & Sarnyai, Z. The Y-maze for assessment of spatial working and reference memory in mice. Methods Mol. Biol. 1916, 105–111 (2019).
Vorhees, C. V. & Williams, M. T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protocols 1, 848–858 (2006).
Acknowledgements
We thank the HCI-CRM Flow Cytometry Core Facility at the Massachusetts General Hospital for assistance in cell sorting; the MGH DF/HCC Specialized Histopathology Services core and the Hope Babette Tang Histology Facility at the Massachusetts Institute of Technology for tissue sectioning and histology services; the MGH NexGen sequencing and Bioinformatics facility for RNA-seq experiments and analysis; L. Wu and the Harvard Genome Modification Facility for help generating mice; G. Wojtkiewicz for help with imaging software; and K. Joyes for copy editing. This work was funded by the Cure Alzheimer’s Fund, the National Institutes of Health (NIH) R35 HL135752, P01 HL131478, P01 HL142494, and the Patricia and Scott Eston MGH Research Scholar (to F.K.S.); NIH K99/R00 HL151750, R01 HL158534, and a Canadian Institutes of Health Research Banting Fellowship (to C.S.M.); HL142494 (to M.N. and B.P.K); NIH R35HL139598 (to M.N.); NIH Ruth L. Kirschstein National Research Service Award Individual Predoctoral Fellowship F31HL147364 (to J.E.M.); and NIH R00 CA218870 (to B.P.K.).
Author information
Authors and Affiliations
Contributions
C.S.M., J.P., A.G., E.K., S.H.C., Y.I., M.G.K., K.A.C., C.V., W.C.P., J.E.M., C.T.C., S.H., H.J., L.P.W., J.D., S.S., A.A., F.K., M.J., and P.F.F. conducted experiments, and collected and analysed data. C.S.M., J.P., A.G., R.I.S., R.W., B.P.K., M.N., R.E.T., and F.K.S. conceptualized and designed experiments, discussed results, and interpreted data. C.S.M. and F.K.S. designed figures and wrote the manuscript. R.E.T. and F.K.S. supervised, directed, and managed the study.
Corresponding authors
Ethics declarations
Competing interests
C.S.M., F.K.S., and R.E.T. are inventors on a patent application filed by Mass General Brigham that describes targeting IL-3 signalling in AD (invention record no. 2020-568). B.P.K. is an inventor on patent applications filed by Mass General Brigham that describe genome engineering technologies and methods, is an advisor to Acrigen Biosciences, and consults for Avectas Inc. and ElevateBio.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Analysis of Il3−/− mice.
a, FITC-dextran (mol. wt. 4000) was injected i.v. into WT and Il3−/− mice before death. Blood–brain barrier integrity was determined by measuring the FITC signal in brain homogenate (n = 6 WT mice; n = 4 Il3−/− mice). b, Before death, WT and Il3−/− mice were injected i.v. with an anti-GR1 antibody conjugated to PE to label all circulating monocytes and neutrophils. PE signal among CD45+ cells was assessed in the brain by flow cytometry (n = 7 WT mice; n = 4 Il3−/− mice). c, Doublecortin staining and quantification in the hippocampus of WT and Il3−/− mice at 4 months of age (n = 3). d, Absence of caspase 3 staining in the hippocampus of WT and Il3−/− mice along with a representative image of rare positively stained cells from the thalamus (n = 3). e, Assessment of MHCII+ microglia in the brains of WT and Il3–/– mice (n = 7 WT mice; n = 4 Il3−/− mice). f, Analysis of Ki67+ proliferating microglia (n = 5). g, Time in new arm during Y-maze testing (n = 7). Groups of mice are of evenly mixed sex. Mean ± s.e.m.
Extended Data Fig. 2 Haematopoiesis and peripheral immune cell dynamics in WT, 5xFAD, and Il3−/−5xFAD mice.
a, Average swim speed during acquisition days of Morris water maze (n = 10 5xFAD mice; n = 9 Il3−/−5xFAD mice). b, Flow cytometry assessment of blood leukocytes (n = 9 WT mice; n = 12 5xFAD mice; n = 8 Il3−/−5xFAD mice). One-way ANOVA. c, Flow cytometry analysis of Lin−SCA1+cKIT+ cells (LSKs), multi-potent progenitors (MPP)-4 and -3, short-term haematopoietic stem cells (StHSCs), long-term HSCs (LtHSCs), common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), and monocyte dendritic progenitors (MDPs) in the bone marrow of 5-month-old WT, 5xFAD, and Il3−/−5xFAD mice (n = 9 WT mice; n = 11 5xFAD mice; n = 8 Il3−/−5xFAD mice). One-way ANOVA. d, FITC-dextran (mol. wt. 4,000) was injected i.v. into 5xFAD and Il3−/−5xFAD mice before death. Blood–brain barrier integrity was determined by measuring the FITC signal in brain homogenates (n = 4). e, WT, 5xFAD, Il3−/−5xFAD mice were joined by parabiosis with UbiGFP mice from the age of 2 to 6 months (4 months total). f, GFP chimerism in blood Ly6Chi monocytes and brain CD45+ cells was assessed by flow cytometry in the WT, 5xFAD, and Il3−/−5xFAD parabionts (n = 4 WT mice; n = 5 5xFAD mice; n = 3 Il3−/−5xFAD mice). Groups of mice are of evenly mixed sex. *P < 0.05, **P < 0.01, ***P < 0.001. Mean ± s.e.m.
Extended Data Fig. 3 Generation and validation of Il3GFPfl/flAldh1l1CreERT25xFAD and Il3rafl/flCx3cr1CreERT25xFAD mice.
a, b, Schematics of the endogenous loci and editing strategies to generate conditional/reporter models for Il3 (a) and Il3ra (b). Mice were generated by excising large fragments of the endogenous loci by co-delivery of SpCas9 and two gRNAs, and in the presence of a long single-stranded DNA donor encoding a loxP-cDNA-P2A-EGFP-loxP cassette. c, d, Representative Sanger sequencing traces validating insertion of the loxP-cDNA-P2A-EGFP-loxP cassettes at the endogenous loci of Il3 (c) and Il3ra (d). Missense mutation quenches the GFP signal in Il3ra-targeted mice. e, GFP signal in ex vivo-stimulated splenic and lymph node T cells, known IL-3 sources, from WT and Il3GFPfl/fl mice. f, Flow cytometry analysis of astrocyte IL-3 production in 5-month-old Il3GFPfl/fl5xFAD and Il3GFPfl/flAldh1l1CreERT25xFAD mice injected with tamoxifen. g, qPCR analysis of Il3 mRNA expression in sorted astrocytes (n = 4 Il3GFPfl/fl5xFAD mice; n = 5 Il3GFPfl/flAldh1l1CreERT25xFAD mice). h, CSF IL-3 levels. Filled circles, males; open circles, females (n = 6 Il3GFPfl/fl5xFAD mice; n = 9 Il3GFPfl/flAldh1l1CreERT25xFAD mice). i, STAT5 phosphorylation in ex vivo heart macrophages from Il3rαfl/fl mice simulated with rIL-3. j, Flow cytometry analysis of microglia IL-3Rα production in 5-month-old Il3rαfl/fl5xFAD and Il3rαfl/flCx3cr1CreERT25xFAD mice injected with tamoxifen. k, qPCR analysis of Il3rα mRNA expression in sorted microglia (n = 4 Il3rαfl/fl5xFAD mice; n = 5 Il3rαfl/flCx3cr1CreERT25xFAD mice). All Il3GFPfl/fl5xFAD, Il3GFPfl/flAldh1l1CreERT25xFAD, Il3rαfl/fl5xFAD, and Il3rαfl/flCx3cr1CreERT25xFAD mice were injected with tamoxifen beginning at 2 months of age. **P < 0.01, ***P < 0.001, two-tailed Mann–Whitney U-tests. Mean ± s.e.m.
Extended Data Fig. 4 Flow cytometry gating strategy and histology controls.
a, Gating strategy used to identify cell populations in the brains of all mice except Aldh1l1GFP mice. b, Backgating of GFP+ astrocytes in Il3GFPfl/fl mice. c, Gating strategy used to identify cell populations in the brain of Aldh1l1GFP mice. d, Gating and isotype control plots for microglia IL-3Rα staining. e, Representative images of IgG control antibody staining and IL-3 staining in the brain.
Extended Data Fig. 5 Astrocyte IL-3 and microglia IL-3Rα production.
a, Il3 expression in astrocytes sorted from WT and 5xFAD mice at 5 and 12 months of age (n = 5 5-month-old WT mice; n = 4 5-month-old 5xFAD mice; n = 3 12-month-old WT mice; n = 4 12-month-old 5xFAD mice). b, Tissue IL-3 levels in various brain regions in WT mice at 4 months of age (n = 4). c, Scheme of daily i.p. LPS injection into WT mice over 4 days and qPCR analysis of C3 and Gfap in sorted astrocytes after LPS injection (n = 4 PBS mice; n = 3 LPS mice for C3; n = 4 LPS mice for Gfap and Il3). Two-tailed Mann–Whitney U-tests. d, CSF IL-3 levels (n = 8 WT PBS mice; n = 6 LPS mice). e, Representative images of GFAP+ astrocytes in the cortex of WT, Il3−/−, 5xFAD, and Il3−/−5xFAD mice. f, Representative images of Aβ deposits (6E10) and astrocytes (GFAP) in 5xFAD and Il3−/−5xFAD mice. g, Proportion of IL-3Rα+ microglia in the brain of WT mice at various ages (n = 4). h, Il3ra transcript expression in brain homogenate from WT mice at various ages (n = 4). i, Proportion of IL-3Rα+ macrophages in heart, liver, lung (interstitial and alveolar), and brain of WT and 5xFAD mice at 8 months of age (n = 3 WT and 5xFAD mice for all tissues except brain where n = 4). One-way ANOVA. *P < 0.05, **P < 0.001. Mean ± s.e.m.
Extended Data Fig. 6 Il3ra expression in microglia and characterization of IL-3Rαhi and IL-3Rαlo microglia.
a, Analysis of Il3ra and other cytokine receptors expressed in homeostatic, DAM stage 1, and DAM stage 2 microglia (data from ref. 23). b, Gating strategy for IL-3Rαhi and IL-3Rαlo microglia from 5xFAD mice. c, Flow cytometry analysis of IL-3Rαhi and IL-3Rαlo microglia (n = 6 except CD11c and CCL2 where n = 3). d, mRNA transcript expression in sorted IL-3Rαhi and IL-3Rαlo microglia (n = 7 for IL-3Rαlo microglia except that n = 6 for Il6, Itgam, Ptprc, Cd36, and Cd68, and n = 5 for Il10; n = 7 for IL-3Rαhi microglia except that n = 6 for Il6, Ptprc and Cd68, and n = 5 for Il10). *P < 0.05, **P < 0.01, two-tailed Mann–Whitney U-tests. Mean ± s.e.m.
Extended Data Fig. 7 IL-3 does not influence proliferation, recognition and phagocytosis of Aβ, or production of inflammatory cytokines in microglia.
a, BrdU incorporation into microglia (n = 6 5xFAD; n = 7 Il3−/−5xFAD). Two-tailed Mann–Whitney U-tests. b, Heat map of microglia RNA-seq expression data for genes important for cell cycle and proliferation. c, Assessment of ex vivo phagocytosis of Aβ42 conjugated to a pH-sensitive dye (PHrodo Red) in sorted microglia lacking Il3 or stimulated with rIL-3 (n = 4). d, Heat map of microglia RNA-seq expression data for genes important for Aβ recognition and phagocytosis. e, Heat map of microglia RNA-seq expression data for inflammatory cytokines. Groups of mice are of evenly mixed sex. Mean ± s.e.m.; ns, not significant.
Extended Data Fig. 8 Human AD iPS triculture system.
a, Representative images demonstrating p-tau (PHF1) localization with neurons (TUJ1) and the presence of astrocytes (GFAP) in the central chamber, and microglia (P2RY12) in the side chamber. b, ELISA quantification of Aβ40, Aβ38, and Aβ42 in medium (n = 3 per group). c, IL3RA expression in human iPS microglia exposed to Aβ (n = 2). d, Chemokine and cytokine levels in the medium of the human AD iPS triculture system (n = 3 except n = 2 for CCL13 AD astrocytes + neurons). Mean ± s.e.m.
Extended Data Fig. 9 rIL-3 delivery to the cortex or periphery of 5xFAD mice and summary figure.
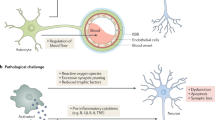
a, Recombinant IL-3 or PBS was delivered into the cortex of 5xFAD mice. Three days later, localization of microglia to Aβ aggregates was assessed (n = 7 PBS mice; n = 6 rIL-3 mice). Two-tailed Mann–Whitney U-tests. b, Scheme of rIL-3 delivery intraperitoneally twice a week for 10 weeks to 5xFAD mice. c, Prior to death, Y-maze behavioural testing was performed and time in the new arm was quantified (n = 7 PBS mice; n = 8 rIL-3 mice). d, The amount of Aβ in the cortex of mice was quantified by analysing histological sections (n = 6). Groups of mice are of evenly mixed sex. **P < 0.01. Mean ± s.e.m. e, Model of the role of IL-3 in AD. Astrocytes produce IL-3. In response to Aβ, TREM2 signalling increases microglia IL-3Rα, rendering microglia responsive to astrocyte-derived IL-3. IL-3 signalling instigates transcriptional and functional programming of microglia, which leads to a signature of immune regulation, motility, and migration. IL-3-dependent programming promotes clustering of microglia around Aβ, which enables clearance of Aβ and mitigation of AD pathology.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5 and Supplementary Sequences 1-2.
Video 1
Three-dimensional confocal imaging of optically-cleared mouse cortex stained for Aβ and microglia.
Rights and permissions
About this article
Cite this article
McAlpine, C.S., Park, J., Griciuc, A. et al. Astrocytic interleukin-3 programs microglia and limits Alzheimer’s disease. Nature 595, 701–706 (2021). https://doi.org/10.1038/s41586-021-03734-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03734-6
This article is cited by
-
Understanding immune microenvironment alterations in the brain to improve the diagnosis and treatment of diverse brain diseases
Cell Communication and Signaling (2024)
-
Taming microglia: the promise of engineered microglia in treating neurological diseases
Journal of Neuroinflammation (2024)
-
Exercise mimetics: a novel strategy to combat neuroinflammation and Alzheimer’s disease
Journal of Neuroinflammation (2024)
-
Osteocyte-derived sclerostin impairs cognitive function during ageing and Alzheimer’s disease progression
Nature Metabolism (2024)
-
Rheumatoid arthritis is a protective factor against Alzheimer’s disease: a bidirectional two-sample Mendelian randomization study
Inflammopharmacology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.