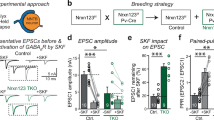
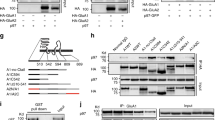
Abstract
Ionotropic glutamate delta receptors 1 (GluD1) and 2 (GluD2) exhibit the molecular architecture of postsynaptic ionotropic glutamate receptors, but assemble into trans-synaptic adhesion complexes by binding to secreted cerebellins that in turn interact with presynaptic neurexins1,2,3,4. It is unclear whether neurexin–cerebellin–GluD1/2 assemblies serve an adhesive synapse-formation function or mediate trans-synaptic signalling. Here we show in hippocampal synapses, that binding of presynaptic neurexin–cerebellin complexes to postsynaptic GluD1 controls glutamate receptor activity without affecting synapse numbers. Specifically, neurexin-1–cerebellin-2 and neurexin-3–cerebellin-2 complexes differentially regulate NMDA (N-methyl-d-aspartate) receptors and AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptors by activating distinct postsynaptic GluD1 effector signals. Of note, minimal GluD1 and GluD2 constructs containing only their N-terminal cerebellin-binding and C-terminal cytoplasmic domains, joined by an unrelated transmembrane region, fully control the levels of NMDA and AMPA receptors. The distinct signalling specificity of presynaptic neurexin-1 and neurexin-35,6 is encoded by their alternatively spliced splice site 4 sequences, whereas the regulatory functions of postsynaptic GluD1 are mediated by conserved cytoplasmic sequence motifs spanning 5–13 residues. Thus, GluDs are signalling molecules that regulate NMDA and AMPA receptors by an unexpected transduction mechanism that bypasses their ionotropic receptor architecture and directly converts extracellular neurexin–cerebellin signals into postsynaptic receptor responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Exact P values are included with the source data. Source data are provided with this paper.
References
Uemura, T. et al. Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 (2010).
Joo, J. Y. et al. Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem. Biophys. Res. Commun. 406, 627–632 (2011).
Matsuda, K. et al. Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science 328, 363–368 (2010).
Yuzaki, M. & Aricescu, A. R. A GluD coming-of-age story. Trends Neurosci. 40, 138–150 (2017).
Dai, J., Aoto, J. & Südhof, T. C. Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 102, 993–1008.e5 (2019).
Aoto, J., Martinelli, D. C., Malenka, R. C., Tabuchi, K. & Südhof, T. C. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154, 75–88 (2013).
Zhu, F., et al. Architecture of the mouse brain synaptome. Neuron 99, 781–799 (2018).
Nusser, Z. Creating diverse synapses from the same molecules. Curr. Opin. Neurobiol. 51, 8–15 (2018).
Huganir, R. L. & Nicoll, R. A. AMPARs and synaptic plasticity: the last 25 years. Neuron 80, 704–717 (2013).
Nakazawa, K. & Sapkota, K. The origin of NMDA receptor hypofunction in schizophrenia. Pharmacol. Ther. 205, 107426 (2020).
Singh, T., Neale, B. M., & Daly, M. J. Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. Preprint at https://doi.org/10.1101/2020.09.18.20192815 (2020)
Südhof, T. C. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171, 745–769 (2017).
Gomez, A. M., Traunmüller, L. & Scheiffele, P. Neurexins: molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 22, 137–151 (2021).
Hu, Z., Xiao, X., Zhang, Z. & Li, M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Mol. Psychiatry 24, 1400–1414 (2019).
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299 (2016).
Zhu, S. & Gouaux, E. Structure and symmetry inform gating principles of ionotropic glutamate receptors. Neuropharmacology 112, 11–15 (2017).
Nakamoto, C. et al. Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain. J. Comp. Neurol. 528, 1003–1027 (2020).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Seigneur, E. & Südhof, T. C. Cerebellins are differentially expressed in selective subsets of neurons throughout the brain. J. Comp. Neurol. 525, 3286–3311 (2017).
Torashima, T. et al. Rescue of abnormal phenotypes in δ2 glutamate receptor-deficient mice by the extracellular N-terminal and intracellular C-terminal domains of the δ2 glutamate receptor. Eur. J. Neurosci. 30, 355–365 (2009).
Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018).
Hirai, H. Ca2+-dependent regulation of synaptic δ2 glutamate receptor density in cultured rat Purkinje neurons. Eur. J. Neurosci. 14, 73–82 (2001).
Sando, R., Jiang, X. & Südhof, T. C. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363, eaav7969 (2019).
Khalaj, A. J. et al. Deorphanizing FAM19A proteins as pan-neurexin ligands with an unusual biosynthetic binding mechanism. J. Cell Biol. 219, e202004164 (2020).
Trotter, J. H. et al. Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. J. Cell Biol. 218, 2677–2698 (2019).
Patzke, C., et al. Neuromodulator signaling bidirectionally controls vesicle numbers in human synapses. Cell 179, 498–513 (2019).
Dai, J., Chen, P., Tian, H. & Sun, J. Spontaneous vesicle release is not tightly coupled to voltage-gated calcium channel-mediated Ca2+ influx and is triggered by a Ca2+ sensor other than synaptotagmin-2 at the juvenile mice calyx of held synapses. J. Neurosci. 35, 9632–9637 (2015).
Jiang, X., Sando, R. & Südhof, T. C. Multiple signaling pathways are essential for synapse formation induced by synaptic adhesion molecules. Proc. Natl Acad. Sci. USA 118, e2000173118 (2021).
Zhang, B. et al. Neuroligins sculpt cerebellar Purkinje-cell circuits by differential control of distinct classes of synapses. Neuron 87, 781–796 (2015).
Zhang, R. S., Liakath-Ali, K. & Südhof, T. C. Latrophilin-2 and latrophilin-3 are redundantly essential for parallel-fiber synapse function in cerebellum. eLife 9, e54443 (2020).
Kakegawa, W. et al. Differential regulation of synaptic plasticity and cerebellar motor learning by the C-terminal PDZ-binding motif of GluRδ2. J. Neurosci. 28, 1460–1468 (2008).
Uemura, T., Mori, H. & Mishina, M. Direct interaction of GluRδ2 with Shank scaffold proteins in cerebellar Purkinje cells. Mol. Cell. Neurosci. 26, 330–341 (2004).
Acknowledgements
We thank L. Chen for advice and the laboratory of H. Hirai for plasmids. This study was supported by a grant from the NIMH (MH052804 to T.C.S.) and fellowships to K.L.-A. from the European Molecular Biology Organization (ALTF 803-2017) and the Larry L. Hillblom Foundation (2020-A-016-FEL).
Author information
Authors and Affiliations
Contributions
J.D. performed all experiments and analysed all data except for the immunoblotting and in situ hybridization experiments, which were performed by C.P. and K. L.-A., respectively. E.S. contributed mouse husbandry and genotyping. J.D. and T.C.S. conceptualized the project, designed the experiments, analysed the data and wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Zoltan Nusser and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CRISPR-mediated deletion and synaptic localization of GluD1 in cultured hippocampal neurons, and the effect on synapse numbers and synaptic function.
a, Quantitative RT–PCR demonstrating robust GluD1 but weak GluD2 expression in cultured hippocampal neurons, providing the rationale for targeting GluD1 over GluD2. b, Experimental strategy for the CRISPR-mediated GluD1 deletion. Hippocampal cultures from Cas9-expressing transgenic18 mice were infected at DIV4-5 with lentiviruses expressing only mCherry or co-expressing mCherry with GluD1-directed sgRNAs, and were analysed at DIV15-17. c, Screening of five sgRNAs for the GluD1 KO efficacy in hippocampal cultures from Cas9-expressing transgenic mice18. GluD1 mRNA levels were analysed by quantitative RT–PCR in neurons as described in a. sgRNA sequences: sgRNA1, 5′-GCCGACTCCATCATCCACAT-3′; sgRNA2, 5′-GTAACGCACTGGCATATCCA-3′; sgRNA3, 5′-GGCCAATAATCCGTTCCAGG-3′; sgRNA4, 5′-GAACGCAGCCAAGGACGACA-3′; sgRNA5, 5′-CTTCGAGGAGAACGCAGCCA-3′. sgRNA1 was used for all subsequent experiments. d, GluD1 deletion using sgRNA1 in hippocampal cultures from Cas9-expressing transgenic mice abolishes most GluD1 protein expression by the time of analysis used in the current experiments. Protein levels were quantified using immunoblots (see Fig. 1b) with fluorescent secondary antibodies. e, Representative images of more than 3 experiments of hippocampal neurons cultured from Cas9-expressing transgenic mice and infected with a control lentivirus or a lentivirus expressing sgRNA1 targeting GluD1 (CRISPR). Neurons were stained for MAP2 and DAPI; the mCherry signal is from the lentiviruses). f, Representative images of hippocampal neurons cultured from Cas9-expressing transgenic mice and infected with a control lentivirus or a lentivirus expressing sgRNA1 targeting GluD1 (CRISPR, used as a negative control for GluD1 staining). Neurons were stained for vGluT1, MAP2 and GluD1 (left and middle panels) or Homer1 (right panels). g, Co-localization quantifications indicate that the GluD1 and the Homer1 signals are both adjacent to vGluT1 signals and are highly correlated with each other (Pearson correlation test). For minimum distance quantification, the range of 0.1-4 μm was analysed for distances between presynaptic vGlut1 and postsynaptic GluD1 or Homer1. h, CRISPR-mediated deletion of GluD1 has no effect on excitatory synapse numbers in hippocampal neurons, independently confirming the experiments shown in Fig. 1 using double labelling for pre- and postsynaptic excitatory synapse markers (vGluT1 and Homer1, respectively). Only puncta positive for both markers were quantified (left, representative images of neurons stained for vGluT1, Homer1 and MAP2; right, summary graphs of the density of double-positive for vGluT1 and Homer1 synaptic puncta). i, CRISPR-mediated GluD1 deletion does not alter EPSC kinetics (left, AMPAR-EPSC rise time (20–80%); middle, AMPAR-EPSC decay constants at −70 mV fitted with double-exponential function; NMDAR-EPSC decay constants at +40 mV fitted with double-exponential function. Data for i and j are from the experiments of Fig. 1g–i. j, k, CRISPR-mediated GluD1 deletion does not alter the coefficient of variation (j; left, representative traces; right, summary graph of coefficient of variation), or the PPR of AMPAR-EPSCs (k; top, sample traces; bottom, summary plots of the PPRs as a function of interstimulus intervals (ISIs)). Data are means ± s.e.m. (number of independent experiments (data points) is indicated in the bars of a–d, regions of interests/independent experiments is listed in h, and number of neurons/ independent experiments is indicated in i–k). Statistical significance was assessed by two-tailed Student’s t-test or two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; for numerical P values, see source data; non-significant comparisons are not indicated).
Extended Data Fig. 2 Effects of the CRISPR-mediated deletion of GluD1 and Cre-mediated deletion of Cbln2 in vivo.
a, b, Experimental strategy for CRISPR-mediated sparse GluD1 deletion in neurons of the subiculum in the hippocampal formation. Mice were bilaterally infected at P21 by stereotactic injections of lentiviruses co-expressing sgRNA directed against GluD1 and mCherry. Mice were sacrificed at P42-P49, and subiculum sections were analysed by acute slice physiology comparing adjacent infected mCherry-positive GluD1-deficient neurons with non-infected control neurons (strategy outline (a); diagram of stereotactic injections (b)). c, Image of a subiculum section from a stereotactically infected mouse (representative of 6 experiments). Arrows identify sparsely infected mCherry-positive neurons. d, CRISPR-mediated GluD1 deletion does not alter the coefficient of variation of AMPAR-EPSCs (left, representative traces; right, summary graph of coefficient of variation). e, CRISPR-mediated GluD1 deletion does not alter the EPSC kinetics (left, EPSC rise time (20–80%); middle, EPSC decay constants at −70 mV fitted with double-exponential function; EPSC decay constants at +40 mV fitted with double-exponential function). Data in this panel and panel d were from the experiments in Fig. 1l. f, g, RNA in situ hybridization analyses reveal that cerebellin-1 (Cbln1) is poorly expressed in the hippocampal formation, cerebellin-2 (Cbln2) is detectable only in subiculum neurons, and cerebellin-4 (Cbln4) is found only in a subset of interneurons (overview of the hippocampal formation hybridized for Cbln1, Cbln2, and Cbln4 mRNAs (f); representative images for each cerebellin (g)). Images are representative of multiple brain sections, and match those obtained by the Allen Brain Institute (https://mouse.brain-map.org). h, Experimental strategy for analysis of conditional Cbln2 deletions in vivo using Cbln2 cKO mice19. Mice were bilaterally infected at P21 by stereotactic injections of AAVs expressing ΔCre-eGFP (control) or Cre-eGFP, and subiculum neurons were analysed in acute slices 2-3 weeks later. In this and all following experiments, ΔCre refers to the expression of a mutant inactive Cre that is otherwise indistinguishable from Cre. i, Conditional Cbln2 deletion from subiculum neurons does not impair excitatory synapse numbers (representative images complementing those shown in Fig. 2a). j, Summary graph of the EPSC success rate during minimal stimulation experiments (Fig. 2d). k, Conditional Cbln2 deletion does not alter the coefficient of variation of AMPAR-EPSCs (left, representative traces; right, summary graph of coefficient of variation). l, Image of an acute slice of the hippocampal formation from a mouse infected with AAVs expressing Cre-eGFP (green), and illustration of the electrophysiological recording configuration. m, Conditional Cbln2 deletion does not alter the EPSC kinetics (left, EPSC rise time (20–80%); middle, EPSC decay constants at −70 mV fitted with double-exponential function; EPSC decay constants at +40 mV fitted with double-exponential function). Data in this panel and panel k were from the experiments shown in Fig. 2c. n, Constitutive Cbln2 deletion does not alter the AMPA/NMDA EPSC ratio at CA3-CA1 Schaffer collateral synapses (left, representative traces; right, summary graph). Both conditional and constitutive Cbln2 deletions cause a massive change in this ratio when analysed in subiculum synapses (see Fig. 2). o, Cumulative probability plots show that the conditional Cbln2 deletion in subiculum significantly shifts the distribution of AMPAR-EPSC amplitudes the right and of NMDAR-EPSC amplitudes to the left, as analysed during minimal stimulation experiments. Data are means ± s.e.m. (number of neurons/mice is indicated in the bars of d, e, j, k, m, n). Statistical significance was assessed by two-tailed Student’s t-test or two sample K-S test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; for exact P values, see source data; non-significant relations are not indicated).
Extended Data Fig. 3 Double deletion of GluD1 and Cbln2 produces the same phenotype as individual deletions of GluD1 and Cbln2 in hippocampal cultures.
a, Experimental strategy. Hippocampal cultures from newborn mice expressing Cas9 on the Cbln2 cKO background (cas9 and Cbln2fl/fl) were infected with lentiviruses and analysed as indicated. Immunoblots on the right show that expression of the GluD1-directed sgRNA suppressed GluD1 protein levels with ΔCre or Cre co-expression (for additional data, please also see Supplementary Fig. 1b). b, Quantifications of GluD1 levels show that CRISPR-mediated deletion of GluD1 is efficient even after Cre-mediated deletion of Cbln2. GluD1 protein levels were measured by quantitative immunoblotting using fluorescent secondary antibodies in neurons of the indicated genotypes. c, Representative images of neurons stained for vGluT1 and MAP2. Merged images and analyses of synaptic properties are shown in Fig. 2h. d, Summary graphs of vGluT1 puncta size and vGluT1 staining intensity as a function of the Cbln2-GluD1 double deletion (additional analyses for Fig. 2h). e, f, Conditional Cbln2 deletion has no effect on excitatory synapse numbers in hippocampal neurons as analysed in independent experiments complementing those of Fig. 2, but using double labelling for a presynaptic (vGluT1) and postsynaptic marker (Homer1). Only double-positive puncta were quantified (representative images of neurons stained for vGluT1, Homer1 and MAP2 (e); summary graph of the density of vGluT1 & Homer1 double-positive synaptic puncta(f)). g, Double GluD1 and Cbln2 deletion does not alter the coefficient of variation of AMPAR-EPSCs (left, representative traces; right, summary graph of coefficient of variation). h, Double GluD1 and Cbln2 deletion does not alter EPSC kinetics (left, EPSC rise time (20–80%); middle, AMPAR-EPSC decay constants at −70 mV fitted with a double-exponential function; right, NMDAR-EPSC decay constants at +40 mV fitted with a double-exponential function). Data in g and h were from the experiments of Fig. 2i–l. i. Measurements of PPRs of evoked AMPAR-EPSCs show that the double GluD1 and Cbln2 deletion does not significantly alter PPRs (left, representative traces; right, summary plot of PPRs as a function of ISIs). Data are means ± s.e.m. (n’s in b, independent experiments (data points) are shown in bars; in d and f, dendrites/independent experiments; in g–i, neurons/independent experiments are listed). Statistical significance was assessed by two-tailed Student’s t-test or two-way ANOVA (**P ≤ 0.01, and ***P ≤ 0.001; for numerical P values, see source data; non-significant comparisons are not indicated).
Extended Data Fig. 4 Mechanism of Nrxn1βSS4+ and Nrxn3βSS4+ regulation of NMDARs and AMPARs.
a, Experimental strategy for testing the effect of the CRISPR-mediated GluD1 deletion on Nrxn1βSS4+- and Nrxn3βSS4+-mediated signalling in hippocampal cultures. Neurons cultured from Cas9 mice were infected at DIV4-5 with lentiviruses expressing only mCherry or co-expressing mCherry and GluD1-sgRNA, and again at DIV 6-7 with lentiviruses expressing various Nrxn1β or Nrxn3β constructs. Neurons were analysed at DIV15-17. b, Nrxn1βSS4-, Nrxn1βSS4+, Nrxn3βSS4-, and Nrxn3βSS4+ are similarly expressed in hippocampal cultures (top, sample blots; bottom, quantification of protein levels using Flag-tag antibodies; for additional data, please also see Supplementary Fig. 1c). c, Quantifications of Nrxn1βSS4-, Nrxn1βSS4+, Nrxn3βSS4-, and Nrxn3βSS4+ surface levels show that all constructs are efficiently transported to the cell-surface (left, representative images; right, summary graphs of the staining intensity of surface Flag-tagged proteins). Neurons were prepared as described in a. d–h, Nrxn1βSS4-, Nrxn1βSS4+, Nrxn3βSS4-, and Nrxn3βSS4+ expression and GluD1 deletions, alone or in combination, did not change the kinetics of AMPAR- and NMDAR-EPSCs or the PPRs of AMPAR-EPSCs (AMPAR-EPSC rise time (20–80%) (d); AMPAR-EPSC decay constants at −70 mV fitted with double-exponential function [top, fast component; bottom, slow component] (e); NMDAR-EPSC decay constants at +40 mV fitted with double-exponential function [top, fast component; bottom, slow component] (f); representative PPR traces (g); summary graphs of PPRs as a function of inter-stimulus intervals (h)). Data are from experiments in Fig. 3. i, Sequences of the alternatively spliced inserts of Nrxn1βSS4+ and Nrxn3βSS4+ and of the ‘swap’ mutants of these alternatively spliced inserts, referred to as Nrxn1βN3-SS4+ and Nrxn3βN1-SS4+, in which the SS4+ sequence of Nrxn1βSS4+ is introduced into Nrxn3βSS4+, and vice versa. j, k, Same as b and c, but with expression of Nrxn1βN3-SS4+ and Nrxn3βN1-SS4+ (for additional data, please also see Supplementary Fig. 1d). l–p, Nrxn1βN3-SS4+ and Nrxn3βN1-SS4+, alone or in combination with the GluD1 KO, do not change the kinetics of AMPAR- and NMDAR-EPSCs or the paired-pulse ratio of AMPAR-EPSCs (AMPAR-EPSC rise time (20–80%) (l); AMPAR-EPSC decay constants at −70 mV fitted with a double-exponential function (m); NMDAR-EPSC decay constants at +40 mV fitted with a double-exponential function (n); representative PPR traces (o); summary graphs of PPRs as a function of inter-stimulus intervals (p)). Data are from experiments in Fig. 3. Data are means ± s.e.m.; independent experiments (data points) are shown in bars (b, j); numbers of sections/independent experiments are shown in bars (c, k); neurons/independent experiments are listed (d–f, h, l–n, p). Please see source data for details.
Extended Data Fig. 5 Design of GluD1 mutant proteins and of minimal GluD1-CD4 and GluD2-CD4 proteins.
a, Alignment of the C-terminal sequences of mouse GluD1 and GluD2 to illustrate the location of cytoplasmic tail mutations in GluD1 and the positions where GluD sequences were fused to the CD4 transmembrane region in the GluD1-CD4TMR and GluD2-CD4TMR constructs (dotted lines indicate sequence not shown; ATD, amino-terminal domain; TMR 3, third transmembrane region that is followed by the cytoplasmic tail). The five highly conserved sequence motifs in the cytoplasmic C-terminal sequence that were mutated in rescue constructs are highlighted and labelled. In mutants #1–#4, the four internal sequence motifs #1–#4 were replaced by the same GQGHG sequences. The consensus sequence for type 1 PDZ domains at the very C terminus of GluD1 and GluD2 (GTSI) was replaced with a consensus sequence for type 2 PDZ domains (EYYV). This replacement blocks the interaction of the C-terminal PDZ-domain binding sequence of GluD1 with type 1 PDZ-domain proteins, an interaction that has a well documented essential function for GluD2 in cerebellar long-term synaptic plasticity31,32. b, List and description of all GluD1 and GluD2 mutants analysed. In the hybrid GluD1-CD4TMR and GluD2-CD4TMR constructs, all GluD1 and GluD2 sequences from the end of the ATD (that is, the beginning of the presumptive glutamate-binding domain) until the end of the last (3rd) TMR (for GluD1, residues 437–851; for GluD2, residues 441–851) were replaced by a linker and the TMR of CD4 (see shown sequences). As a result, GluD-CD4TMR hybrid proteins lack the TMRs and presumptive glutamate-binding domain of GluDs. In addition, GluD1-CD4TMR and GluD2-CD4TMR contain a shortened cytoplasmic tail with only 59 GluD1 residues, lacking the juxta-membranous part of the cytoplasmic tail. The minimal GluD1-CD4TMR constructs contain only short sequence motifs (5–13 residues) that are mutated in mutants #3 or #4.
Extended Data Fig. 6 GluD1 mutants and GluD1-CD4TMR and GluD2-CD4TMR constructs express well on the neuronal cell surface.
a, Experimental strategy for rescue experiments with GluD1 and GluD2 rescue constructs in GluD1-deficient in hippocampal cultures from Cas9 mice. Cells were infected at DIV4-5 with lentiviruses expressing mCherry only or mCherry together with the GluD1 sgRNA, and again at DIV 6-7 with lentiviruses expressing the wild-type and mutant GluD1 and GluD2 constructs. Neurons were analysed at DIV15-17. b–d, Most mutant GluD1 and GluD2 proteins are similarly expressed in hippocampal cultures (representative immunoblots (for additional data, please also see Supplementary Fig. 1e–g) (b, c); quantification of GluD1 and GluD2 expression using GluD1 antibodies [top] or HA-antibodies recognizing the HA-epitope tag that was added to the N terminus of the rescue proteins [bottom] (d)). Note that although some constructs do not express as well as others (for example, mutant #5), all constructs were overexpressed compared to endogenous GluD1 protein levels. e, f, Quantifications of the surface levels of GluD1 and GluD2 rescue proteins reveal efficient transport of all proteins to the cell-surface. Surface exposure was measured using staining of non-permeabilized fixed neurons with antibodies to extracellular epitopes (representative images (e); summary graphs of the staining intensity of internal HA-tagged proteins (top of panel), of surface HA-tagged proteins (middle of panel) and the ratio of surface/total proteins (bottom of panel) (f)). Neurons were stained first for surface HA before permeabilization, and then for internal HA after permeabilization using different HA antibodies. Data are means ± s.e.m. (independent experiments (data points) are shown in bars for d, sections/independent experiments are shown in the bars for f. Please see source data for details.
Extended Data Fig. 7 Analysis of EPSC kinetics and PPRs in various rescue experiments with cultured GluD1-deficient neurons.
a–i, Quantification of EPSC kinetics for measurements of evoked EPSCs under various GluD1 KO and rescue conditions as indicated, demonstrating that the EPSC kinetics do not change with any experimental manipulation (EPSC rise time (20–80%) (a, d, g); AMPAR-EPSC decay constants recorded at −70 mV and fitted with a double-exponential function (top, fast component; bottom, slow component (b, e, h); NMDAR-EPSC decay constants recorded at +40 mV and fitted with a double-exponential function (top, fast component; bottom, slow component (c, f, i)). Data are from the experiments described in Figs. 4c–e, n–p, 5c–e. j, Representative traces of PPR recordings in cultured neurons under the indicated conditions. k, Summary graphs of the PPRs as a function of the inter-stimulus intervals under the indicated conditions. Data are means ± s.e.m. (for a–i, n values are shown in Figs. 4c–e, n–p, 5c–e; for j, k, neurons/independent experiments are shown in k). Statistical significance was assessed by two-tailed Student’s t-test or two-way ANOVA. No statistically significant differences were observed. Please see source data for details.
Extended Data Fig. 8 Analysis of wild-type and mutant GluD1N-CD4TMR-GluD935–993 proteins.
a, Design of wild-type and mutant GluD1N-CD4TMR-GluD935–993 constructs, corresponding to the GluD1-CD4TMR construct (Fig. 4l) with a greatly shortened C-terminal tail. In the Mut3 and Mut4 constructs, the internal sequence motifs were replaced by the same ‘GQGHG’ sequence, and the C-terminal ‘GTSI’ sequence was mutated to ‘EYYV’ (Extended Data Fig. 5). b, c, Immunoblotting analyses show that most mutant GluD1 proteins are similarly expressed in hippocampal cultures (representative immunoblots (b); quantification of GluD1 expression using the HA-epitope tag that was added to the N terminus of the rescue proteins (c); for additional data, please also see Supplementary Fig. 1h). d, e, Quantifications of GluD1 rescue protein surface levels show that wild-type and Mut3- and Mut4-mutant GluD1N-CD4TMR-GluD935–993 proteins are efficiently transported to the cell-surface (representative images (d); summary graphs of the staining intensity of internal HA-tagged proteins (left), of surface HA-tagged proteins (middle) and the ratio of surface/total proteins (right) (e)). Surface exposure was measured by staining neurons first for surface HA before permeabilization, and then for internal HA after permeabilization using different HA antibodies. f, Representative traces of evoked AMPAR- and NMDAR-EPSCs recorded in hippocampal cultures without or with the GluD1 deletion and rescue with WT, Mut3, and Mut4 GluD1N-CD4TMR-GluD1935–993 constructs. g, Summary graphs of the AMPA/NMDA ratio illustrating that the GluD1 deletion enhances the AMPA/NMDA ratio ~300%, that the WT GluD1N-CD4TMR-GluD1935–993 construct fully rescues this phenotype, but that Mut3- and Mut4-mutant GluD1N-CD4TMR-GluD1935–993 constructs only partly rescue. Neurons were infected with lentiviruses encoding mCherry without or with the GluD1 sgRNA at DIV4-5, re-infected with lentiviruses encoding various GluD1 constructs at DIV6-7, and analysed at DIV15-17. h, i, Summary graphs of the amplitudes of AMPAR-EPSCs (h) and NMDAR-EPSCs (i) demonstrating that the GluD1 deletion increases AMPARs (~150%) but decreases NMDARs (~40%), and that WT GluD1N-CD4TMR-GluD1935–993 fully rescues both phenotypes, whereas Mut3-mutant GluD1N-CD4TMR-GluD1935–993 rescues only the AMPAR but not the NMDAR phenotype and Mut4-mutant GluD1N-CD4TMR-GluD1935–993 rescues only the NMDAR but not the AMPAR phenotype. j, None of the GluD1 manipulations affects the PPR of AMPAR-EPSCs (left, representative traces of PPR recordings; right, summary graphs of PPRs as a function of the inter-stimulus intervals). Data are means ± s.e.m. (n values are shown in bars: independent experiments (data points) (c); sections/independent experiments (e); neurons/independent experiments (g, j)). Statistical significance was assessed by two-tailed Student’s t-test or two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; for numerical P values, see source data; non-significant comparisons are not indicated).
Extended Data Fig. 9 Validation of minimal GluD1-CD4TMR proteins containing only short cytoplasmic GluD1 sequence motifs.
a, b, Minimal GluD1-CD4TMR proteins containing only short cytoplasmic GluD1 sequence motifs proteins are expressed at similar levels in cultured hippocampal neurons (representative immunoblots (a); quantification of GluD1 expression using the N-terminal HA-epitope tag of rescue proteins (b); for additional data, please also see Supplementary Fig. 1i). See Extended Data Fig. 5 for an explanation of the constructs used. c, Surface quantifications show that all minimal GluD1-CD4TMR proteins are efficiently transported to the cell-surface (left, representative images; right, summary graphs of the staining intensity of internal HA-tagged proteins (top), of surface HA-tagged proteins (middle) and of the ratio of surface/total proteins (bottom panel)). Surface exposure was measured by staining neurons first for surface HA before permeabilization, and then for internal HA after permeabilization using different HA antibodies. Controls demonstrated that fixed non-permeabilized cells are impermeant to antibodies. d–f, Summary graphs demonstrating that minimal GluD1-CD4TMR proteins do not change the NMDAR- or AMPAR-EPSC kinetics (AMPAR-EPSC rise times (20–80%) (d); AMPAR-EPSC decay constants at −70 mV fitted with a double-exponential function (top, fast component; bottom, slow component) (e); NMDAR-EPSC decay constants at +40 mV fitted with a double-exponential function (top, fast component; bottom, slow component) (f)). Data are from the experiments in Fig. 5g–i. g, h, The GluD1 deletion without or with expression of minimal GluD1-CD4TMR rescue constructs does not change the paired-pulse ratio (PPR) of AMPAR-EPSCs (representative traces (g); summary graphs of PPRs as a function of the inter-stimulus intervals under the indicated conditions (h)). Data are means ± s.e.m. (number in bars: independent experiments (data points) (b); sections/independent experiments (c); neurons/independent experiments (h); numbers of cells/independent experiments for d–f are given in Fig. 5). Statistical significance was assessed by two-tailed Student’s t-test or two-way ANOVA. No statistically significant differences were observed. Please see source data for details.
Extended Data Fig. 10 Mechanistic model of Nrxn1SS4+→Cbln2→GluD1→NMDAR and Nrxn3SS4+→Cbln2→GluD1→AMPAR signalling pathways.
The model, based on the present data, proposes that Nrxn1- and Nrxn3-alternative splicing at SS4 regulates the postsynaptic content of AMPARs and NMDARs at hippocampal synapses in the subiculum but not the CA1 region by binding to Cbln2, which in turn binds to GluD1 (see Figs. 1 and 2). Surprisingly, binding to the same GluD1-Cbln2 complex enables Nrxn1SS4+ to enhance NMDARs but not AMPARs, whereas binding of Nrxn3SS4+ to the GluD1-Cbln2 complex suppresses AMPARs but not NMDARs (see Fig. 3). Signal transduction by the Nrxn1SS4+-Cbln2-GluD1 and Nrxn3SS4+-Cbln2-GluD1 assemblies is mediated by distinct cytoplasmic GluD1 sequences (HRSPN for AMPAR regulation by Nrxn3SS4+, SRTLS for NMDAR regulation by Nrxn1SS4+), suggesting that GluD1 couples Nrxn1SS4+ and Nrxn3SS4+ signals to distinct effector sequences (see Figs. 4 and 5). Since GluD1 and GluD2 constructs lacking the native GluD1 transmembrane architecture are fully active (see Fig. 4 and 5), binding of Nrxn1SS4+-Cbln2 and Nrxn3SS4+-Cbln2 complexes to the ATD of GluD1 (and of GluD2) activates a transmembrane signal without inducing a conformational change in the transmembrane regions of GluDs.
Supplementary information
Supplementary Figure
Original full-sized immunoblots for all protein level analyses.
Source data
Rights and permissions
About this article
Cite this article
Dai, J., Patzke, C., Liakath-Ali, K. et al. GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature 595, 261–265 (2021). https://doi.org/10.1038/s41586-021-03661-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03661-6
This article is cited by
-
Glutamine metabolism-related genes and immunotherapy in nonspecific orbital inflammation were validated using bioinformatics and machine learning
BMC Genomics (2024)
-
The hidden face of GluD1 at inhibitory synapses
Cell Research (2024)
-
GRID1/GluD1 homozygous variants linked to intellectual disability and spastic paraplegia impair mGlu1/5 receptor signaling and excitatory synapses
Molecular Psychiatry (2024)
-
A combinatorial code of neurexin-3 alternative splicing controls inhibitory synapses via a trans-synaptic dystroglycan signaling loop
Nature Communications (2023)
-
Neurexin-3 subsynaptic densities are spatially distinct from Neurexin-1 and essential for excitatory synapse nanoscale organization in the hippocampus
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.