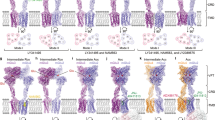
Abstract
The metabotropic glutamate receptors (mGlus) are involved in the modulation of synaptic transmission and neuronal excitability in the central nervous system1. These receptors probably exist as both homo- and heterodimers that have unique pharmacological and functional properties2,3,4. Here we report four cryo-electron microscopy structures of the human mGlu subtypes mGlu2 and mGlu7, including inactive mGlu2 and mGlu7 homodimers; mGlu2 homodimer bound to an agonist and a positive allosteric modulator; and inactive mGlu2–mGlu7 heterodimer. We observed a subtype-dependent dimerization mode for these mGlus, as a unique dimer interface that is mediated by helix IV (and that is important for limiting receptor activity) exists only in the inactive mGlu2 structure. The structures provide molecular details of the inter- and intra-subunit conformational changes that are required for receptor activation, which distinguish class C G-protein-coupled receptors from those in classes A and B. Furthermore, our structure and functional studies of the mGlu2–mGlu7 heterodimer suggest that the mGlu7 subunit has a dominant role in controlling dimeric association and G-protein activation in the heterodimer. These insights into mGlu homo- and heterodimers highlight the complex landscape of mGlu dimerization and activation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Atomic coordinates and cryo-EM density maps for the structures of inactive mGlu2, agonist- and PAM-bound mGlu2, inactive mGlu7 and inactive mGlu2–mGlu7 have been deposited in the PDB under accession codes 7EPA, 7EPB, 7EPC and 7EPD, respectively, and in the Electron Microscopy Data Bank under accession codes EMD-31235, EMD-31236, EMD-31237 and EMD-31238, respectively. Atomic coordinates for the structures of mGlu2 TMD–NAM563 and mGlu2 TMD–NAM597 have been deposited in the PDB under accession codes 7EPE and 7EPF, respectively. The uncropped gels shown in Extended Data Fig. 1r, u, aa are displayed in Supplementary Fig. 1. Any other relevant data are available from the corresponding authors upon reasonable request.
References
Niswender, C. M. & Conn, P. J. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 (2010).
Levitz, J. et al. Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron 92, 143–159 (2016).
Kniazeff, J., Prézeau, L., Rondard, P., Pin, J. P. & Goudet, C. Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol. Ther. 130, 9–25 (2011).
Pin, J. P. & Bettler, B. Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68 (2016).
Bungard, C. J. et al. Quinoline carboxamide and quinoline carbonitrile derivatives as mGluR2-negative allosteric modulators, compositions, and their use. International Patent Application WO 2013/066736 A1 (2013).
Suzuki, G. et al. In vitro pharmacological characterization of novel isoxazolopyridone derivatives as allosteric metabotropic glutamate receptor 7 antagonists. J. Pharmacol. Exp. Ther. 323, 147–156 (2007).
Monn, J. A. et al. Design, synthesis, and pharmacological characterization of (+)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY354740): a potent, selective, and orally active group 2 metabotropic glutamate receptor agonist possessing anticonvulsant and anxiolytic properties. J. Med. Chem. 40, 528–537 (1997).
Cid, J. M. et al. Discovery of 1-butyl-3-chloro-4-(4-phenyl-1-piperidinyl)-(1H)-pyridone (JNJ-40411813): a novel positive allosteric modulator of the metabotropic glutamate 2 receptor. J. Med. Chem. 57, 6495–6512 (2014).
Scholler, P. et al. Allosteric nanobodies uncover a role of hippocampal mGlu2 receptor homodimers in contextual fear consolidation. Nat. Commun. 8, 1967 (2017).
Arasappan, A. et al. Tetrahydronaphthyridine derivatives as mGluR2-negative allosteric modulators, compositions, and their use. International Patent Application WO 2016/029454 A1 (2016).
Koehl, A. et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019).
Chappell, M. D. et al. Discovery of (1S,2R,3S,4S,5R,6R)-2-amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid hydrochloride (LY3020371·HCl): a potent, metabotropic glutamate 2/3 receptor antagonist with antidepressant-like activity. J. Med. Chem. 59, 10974–10993 (2016).
Xue, L. et al. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 11, 134–140 (2015).
Gutzeit, V. A. et al. Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor. eLife 8, e45116 (2019).
Huang, S. et al. Interdomain movements in metabotropic glutamate receptor activation. Proc. Natl Acad. Sci. USA 108, 15480–15485 (2011).
Hlavackova, V. et al. Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci. Signal. 5, ra59 (2012).
Lin, S. et al. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature https://doi.org/10.1038/s41586-021-03495-2 (2021).
Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014).
Doré, A. S. et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014).
Habrian, C. H. et al. Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat. Commun. 10, 5572 (2019).
Liu, J. et al. Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer. eLife 6, e26985 (2017).
Saugstad, J. A., Kinzie, J. M., Mulvihill, E. R., Segerson, T. P. & Westbrook, G. L. Cloning and expression of a new member of the l-2-amino-4-phosphonobutyric acid-sensitive class of metabotropic glutamate receptors. Mol. Pharmacol. 45, 367–372 (1994).
Francesconi, A. & Duvoisin, R. M. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J. Biol. Chem. 273, 5615–5624 (1998).
Olsen, R. H. J. et al. TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat. Chem. Biol. 16, 841–849 (2020).
Fegan, A., White, B., Carlson, J. C. & Wagner, C. R. Chemically controlled protein assembly: techniques and applications. Chem. Rev. 110, 3315–3336 (2010).
Huang, X. et al. Amorphous nickel titanium alloy film: a new choice for cryo electron microscopy sample preparation. Prog. Biophys. Mol. Biol. 156, 3–13 (2020).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Ramlaul, K., Palmer, C. M., Nakane, T. & Aylett, C. H. S. Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. J. Struct. Biol. 211, 107545 (2020).
Kaur, S. et al. Local computational methods to improve the interpretability and analysis of cryo-EM maps. Nat. Commun. 12, 1240 (2021).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 (2012).
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protocols 4, 706–731 (2009).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012).
Jalan-Sakrikar, N. et al. Identification of positive allosteric modulators VU0155094 (ML397) and VU0422288 (ML396) reveals new insights into the biology of metabotropic glutamate receptor 7. ACS Chem. Neurosci. 5, 1221–1237 (2014).
Acknowledgements
The cryo-EM studies were performed at the Center for Biological Imaging (CBI) (http://cbi.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences and the electron microscopy facility of Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences. We thank B. Zhu, X. Huang, X. Li, L. Chen and X. Niu from CBI and Q. Wang from SIMM for their help with cryo-EM data collection. The synchrotron radiation experiments were performed at the BL41XU of SPring-8 with approval of the Japan Synchrotron Radiation Research Institute (proposal no. 2019A2543, 2019B2543, 2019A2541 and 2019B2541). We thank the beamline staff members K. Hasegawa, N. Mizuno, T. Kawamura and H. Murakami of the BL41XU for help with X-ray data collection. We thank Merck for providing the mGlu2 NAMs NAM563 and NAM597. This work was supported by the National Key R&D Program of China 2018YFA0507000 (Q.Z., B.W. and J.L.), National Science Foundation of China grants 31825010 (B.W.), 31830020 (F.S.) and 81720108031 (J.L.), CAS Strategic Priority Research Programs XDB37030100 (Q.Z. and B.W.) and XDB37040102 (F.S.), Agence Nationale de la Recherche, ANR 18-CE11-0004-01 and Fondation Recherche Médicale (DEQ20170336747) (J.-P.P.), and ANR-15-CE18-0020-01, CNRS PRC no. 1403 and Inserm IRP BrainSignal (P.R.).
Author information
Authors and Affiliations
Contributions
J.D. developed the protein expression and purification procedures of mGlu2, prepared the mGlu2 protein samples for cryo-EM and crystallization, solved the crystal structures of mGlu2 TMD–NAMs, performed the IP accumulation assay and helped with manuscript preparation. D.W. developed the protein expression and purification procedures of mGlu7, prepared the mGlu7 protein sample for cryo-EM, performed model building and structure refinement of the homodimers, and helped with the IP accumulation assay and manuscript preparation. H.F. performed the cryo-EM studies of the mGlu2 homodimers. C.X. performed crosslinking, calcium release and FRET assays. L.T. performed the cryo-EM studies of the mGlu7 homodimer. S.L. developed the protein expression and purification procedures of the mGlu2–mGlu7 heterodimer, prepared the mGlu2–mGlu7 protein sample for cryo-EM, and performed model building and structure refinement of the heterodimer. S.H. performed the cryo-EM studies of the mGlu2–mGlu7 heterodimer. Q.T. performed the BRET assay. Xinwei Wang and T.X. helped with protein preparation and functional assays. H.Z. collected the crystal diffraction data. X.C. and C.Y. expressed the proteins. P.L. and Xiaomei Wang helped with crosslinking and calcium release assays. J.-P.P. and P.R. helped with data analysis and interpretation, and edited the manuscript. Y.Z. and H.L. provided mGlu2 ligands for structural and functional studies. J.L. oversaw crosslinking, calcium release and FRET assays, helped with data analysis and interpretation, and edited the manuscript. F.S. oversaw the cryo-EM studies of mGlu2 and mGlu7 homodimers, helped with data analysis and interpretation, and edited the manuscript. B.W. and Q.Z. initiated the project, planned and analysed experiments, supervised the research and wrote the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Karen J. Gregory, Tracy Josephs and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Optimization and characterization of mGlu2 and mGlu7 homo- and heterodimers.
a, Comparison of the mGlu2 homodimers with different C-terminal truncations. The curves of size-exclusion chromatography (SEC) of purified protein samples show higher yield and better homogeneity for the protein with the C-terminal residues V826–L872 truncated. b, Comparison of the mGlu7 homodimers with different C-terminal truncations. The SEC curves of purified protein samples show higher yield and better homogeneity for the protein with the C-terminal residues K860–I915 truncated. c, Comparison of the mGlu2 homodimers with different mutations. The SEC curves of purified protein samples show higher yield for the mutant N6553.52Y/H8157.53Y (construct 1, used to determine the inactive mGlu2 structure). d, Comparison of the mGlu7 homodimers with different mutations. The SEC results of purified protein samples show higher yield for the mutant N6783.52Y/G7224.52I/I7755.59F/P7896.38Y (construct 2, used to determine the inactive mGlu7 structure). e, LY354740-induced IP accumulation assay of wild-type (WT) mGlu2 and mutants. The IP accumulation data are mean ± s.e.m. from at least three independent experiments performed in technical triplicate (same for below). The numbers of independent experiments (n) are shown in the parentheses (same for below). Surface expression levels of the mutants are reported as per cent compared to the wild type from at least three independent measurements performed in duplicate (% of WT): N6553.52Y, 112 ± 29; H8157.53Y, 56 ± 10; S601A, 88 ± 13; C-truncation (V826–L872), 53 ± 3. f, Inhibition of LY354740-induced IP accumulation of wild-type mGlu2 and mutants by NAM563. g, Glutamate-induced IP accumulation assay of wild-type mGlu7 and mutants in the presence of VU0422288. Surface expression levels of the mutants (% of WT): N6783.52Y, 200 ± 25; G7224.52I, 144 ± 14; I7755.59F, 136 ± 13; P7896.38Y, 129 ± 12; C-truncation (K860–I915), 110 ± 10. h, Inhibition of glutamate-induced IP accumulation of wild-type mGlu7 and mutants by MMPIP. i, LY354740-induced Gi activation of wild-type mGlu2 and mutants measured by the BRET assay. The BRET data are mean ± s.e.m. from at least three independent experiments performed in technical duplicate (same for below). j, Glutamate-induced Gi activation of wild-type mGlu7 and mutants in the presence of VU0422288 measured by the BRET assay. k, Ligand screening for the inactive mGlu2 homodimer. The SEC curves show that the NAM563-bound mGlu2 has higher yield and better homogeneity compared to the apo receptor and LY341495-bound mGlu2. l, Ligand screening for the inactive mGlu7 homodimer. The SEC curves show that the NAM MMPIP-bound mGlu7 has higher yield and better homogeneity compared to the apo receptor and the receptor bound to other ligands. m, Inhibition of LY354740-induced IP accumulation of wild-type mGlu2 by NAM563 or NAM597. n, Comparison of wild-type mGlu2 and construct 3 (used to determine the agonist- and PAM-bound mGlu2 structure). The SEC curves of purified protein samples show higher yield and better homogeneity for construct 3. o, JNJ-40411813-induced IP accumulation assay of wild-type mGlu2 and mutants. p, JNJ-40411813-induced Gi activation of wild-type mGlu2 and mutants measured by the BRET assay. q, Ligand screening for the agonist- and PAM-bound mGlu2 homodimer. The SEC curves show higher yield for the agonist- or PAM-bound receptor than the apo protein. r, Nu-PAGE and western blot results of the mGlu2–mGlu7 heterodimer. Double staining (anti-His for His-tagged mGlu2 and anti-Strep for Strep-tagged mGlu7) confirms the existence of mGlu2–mGlu7 heterodimer. Three independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1. s, t, Glutamate-induced IP accumulation of wild-type mGlu2 and mutants. Extended Data Table 3 provides detailed independent experiment numbers (n), statistical evaluation and expression level. u, Crosslinking of the mGlu2 mutants C121A/L6984.51C, C121A/V6994.52C and C121A/V7826.59C–C121A/V7897.27C with (+) or without (−) preincubation with the agonist LY354740 and PAM JNJ-40411813 or the antagonist LY341495 and NAM Ro64-5299. The mutant C121A/V7826.59C–C121A/V7897.27C was tested using an optimized GABAB quality control system, in which the C-terminal tails of the two mGlu2 subunits were replaced by the modified C1 and C2 domains of GABAB1 and GABAB2, respectively, to only allow the mGlu2 dimers containing the C1–C2 dimer to reach the cell surface15. The wild-type and mutant C121A were tested in parallel as controls. Three independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1. v, LY354740-induced IP accumulation assay of mGlu2 in the presence of the oxidant CuP. Surface expression levels of the mGlu2 mutants are reported as per cent compared to the wild type from at least three independent measurements performed in duplicate (% of WT): C121A, 100 ± 6; C121A/L6984.51C, 84 ± 11; C121A/V6994.52C, 92 ± 9. w, LY354740-induced IP accumulation assay of the mGlu2 mutants in the CRD. Extended Data Table 3 provides detailed statistical evaluation and expression level. x, Glutamate-induced IP accumulation assay of wild-type mGlu7 and mutants in the presence of VU0422288. Extended Data Table 3 provides detailed statistical evaluation and expression level. y, LY354740-induced IP accumulation assay of mGlu2 mutants in the CRD and ECL2. Extended Data Table 3 provides detailed independent experiment numbers (n), statistical evaluation and expression level. z, JNJ-40411813-induced IP accumulation assay of wild-type mGlu2 and the mutant in ECL2. aa, Crosslinking of the mGlu2–mGlu7 mutants C121A/D174C (mGlu2)–C136A/R191C (mGlu7) and C121A/S176C (mGlu2)–C136A/R191C (mGlu7). The crosslinking studies were carried out using the GABAB C1–C2 quality control system that allows cell surface targeting of the heterodimer only21. The wild-type heterodimer and mutant C121A (mGlu2)–C136A (mGlu7) were tested in parallel as controls. Four independent experiments were performed with similar results. Results from a representative experiment are shown. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM processing and 3D reconstruction workflow.
a–f, Results of the inactive mGlu2 homodimer. a, Processing workflow. b, Representative cryo-EM image from four independent experiments with similar results. c, Two-dimensional averages. d, Cryo-EM map coloured according to local resolution (in Å). e, Gold-standard Fourier shell correlation (FSC) curve showing an overall resolution at 3.6 Å. f, Cross-validation of model to cryo-EM density map. FSC curves for the final model versus the final map and half maps are shown in black, red and green, respectively. g–l, Results of the inactive mGlu7 homodimer. g, Processing workflow. h, Representative cryo-EM image from five independent experiments with similar results. i, Two-dimensional averages. j, Cryo-EM map coloured according to local resolution (in Å). k, Gold-standard FSC curves showing an overall resolution at 4.0 Å and a resolution at 3.6 Å for the extracellular domains (ECDs). l, Cross-validation of model to cryo-EM density map. m–r, Results of the agonist- and PAM-bound mGlu2 homodimer. m, Processing workflow. n, Representative cryo-EM image from three independent experiments with similar results. o, Two-dimensional averages. p, Cryo-EM map coloured according to local resolution (in Å). q, Gold-standard FSC curve showing an overall resolution at 3.1 Å. r, Cross-validation of model to cryo-EM density map. s–x, Results of the inactive mGlu2–mGlu7 heterodimer. s, Processing workflow. t, Representative cryo-EM image from four independent experiments with similar results. u, Two-dimensional averages. v, Cryo-EM map coloured according to local resolution (in Å). w, Gold-standard FSC curves showing an overall resolution at 3.9 Å and a resolution at 3.5 Å for the extracellular domains. x, Cross-validation of model to cryo-EM density map.
Extended Data Fig. 3 Cryo-EM density maps of mGlu2 and mGlu7 homo- and heterodimers.
The models are shown as cartoon and sticks. The density maps are coloured grey. a, Map and model of the inactive mGlu2 structure shown for all transmembrane helices, B and C helices in VFTs, CRDs and inter-subunit disulfide bond. The model is coloured blue (molecule A) and grey (molecule B). b, Map and model of the inactive mGlu7 structure shown for all transmembrane helices, B and C helices in VFTs, CRDs and inter-subunit disulfide bond. The model is coloured orange (molecule A) and yellow (molecule B). c, Map and model of the agonist- and PAM-bound mGlu2 structure shown for all transmembrane helices, B and C helices in VFTs, LY354740, DN13, CRDs and inter-subunit disulfide bond. The model is coloured blue (molecule A) and grey (molecule B). d, Map and model of the inactive mGlu2–mGlu7 structure shown for all transmembrane helices, B and C helices in VFTs, CRDs, inter-subunit disulfide bond and some non-conserved residues in the two subunits that aided modelling. The model is coloured blue (mGlu2) and orange (mGlu7).
Extended Data Fig. 4 VFT and CRD conformations in mGlus.
a, Comparison of inter-subunit disulfide bonds in the inactive mGlu2 and mGlu7 homo- and heterodimers. The structures of inactive mGlu2 homodimer, mGlu7 homodimer and mGlu2–mGlu7 heterodimer are shown in cartoon representation. The disulfide bonds are shown as yellow sticks. The inter-subunit disulfide bonds are highlighted by a red dashed box. b, Comparison of inter-subunit disulfide bonds in the inactive mGlu2 and agonist- and PAM-bound mGlu2. The structures of inactive mGlu2 homodimer and agonist- and PAM-bound mGlu2 homodimer are shown in cartoon representation. c, Comparison of VFT conformations in inactive mGlu structures. The structures of inactive mGlu2, inactive mGlu5 (PDB code 6N52), inactive mGlu7, apo mGlu2 VFT (PDB code 5KZN) and LY3020371 (antagonist)-bound mGlu2 VFT (PDB code 5KZQ) are shown in cartoon representation. d, Comparison of the VFTs in the inactive structures of mGlu2, mGlu5 and mGlu7. The structures are shown in a side view. The Cα atoms of the mGlu2 residue C234 and its counterparts in mGlu5 and mGlu7 are shown as spheres. The green arrow indicates the shift of the VFT in the mGlu5 structure compared to that in the mGlu2 and mGlu7 structures. e, Comparison of the CRDs in the inactive structures of mGlu2, mGlu5 and mGlu7. The structures are shown in both side and extracellular views. The Cα atoms of the mGlu2 residue C540 and its counterparts in mGlu5 and mGlu7 are shown as spheres. The distance between the Cα atoms of the cysteine residues in each structure is indicated by a dashed line and coloured blue (mGlu2), magenta (mGlu5) or orange (mGlu7). The red arrow indicates the movement of the CRDs in mGlu5 and mGlu7 relative to that in mGlu2. f, Comparison of VFT conformations in agonist-bound mGlu structures. The structures of LY354740- and JNJ-40411813-bound mGlu2, l-quisqualate- and CDPPB-bound mGlu5 (PDB code 6N51), LY354740-bound mGlu2 VFT (PDB code 4XAQ) and glutamate-bound mGlu2 VFT (PDB code 5CNI) are shown in cartoon representation and coloured blue, magenta, red and light gold, respectively. g, Comparison of the CRDs in the agonist- and PAM-bound mGlu2 and mGlu5 structures. The structures are shown in both side and extracellular views. h, Interactions between the CRDs in the agonist- and PAM-bound mGlu2 structure. The residues that are involved in dimerization are shown as blue and grey sticks, respectively. The disulfide bonds are shown as yellow sticks. i, Comparison of VFT conformations in the inactive mGlu2 and mGlu7 homo- and heterodimers. j, VFT dimer interface in the inactive mGlu2–mGlu7 heterodimer structure. The residues that are involved in dimerization are shown as sticks.
Extended Data Fig. 5 Sequence alignment of the TMDs of human mGlus.
Colours represent the similarity of residues: red background, identical; red text, strongly similar. The red arrows indicate the positions with mutations in the inactive mGlu2 and mGlu7 structures (2, mGlu2; 7, mGlu7). The mGlu2 residues that are involved in TMD dimerization in the inactive mGlu2 structure are indicated by green arrows. The conserved residues with the modified Ballesteros–Weinstein numbers for class C GPCRs 1.50, 2.50, 3.50, 4.50, 5.50, 6.50 and 7.50 are indicated by black arrows. The alignment was generated using UniProt (http://www.uniprot.org/align/) and the graphic was prepared on the ESPript 3.0 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Extended Data Fig. 6 Inter- and intra-subunit interactions upon mGlu2 activation and NAM-binding sites in mGlus.
a, Interactions between the CRD and ECL2 in the agonist- and PAM-bound mGlu2 and mGlu5 structures. The structures of agonist- and PAM-bound mGlu2 and mGlu5 (PDB code 6N51) are shown in cartoon representation and coloured blue and pink, respectively. The residues that potentially form interactions between the CRD and ECL2 are shown as sticks. b, Comparison of the helical bundles in the agonist- and PAM-bound structures of mGlu2 and mGlu5. The mGlu2 residues V7826.59 and V7897.27 that are involved in dimerization are shown as sticks. The red arrows indicate the movement of each helix in the mGlu2 structure relative to the mGlu5 structure. c, Crystal structures of mGlu2 TMD–NAMs. The receptor is coloured cyan. The NAMs NAM563 and NAM597 are shown as sticks and coloured yellow and magenta, respectively. The mGlu2 residues that form interactions with the NAMs are shown as cyan sticks (middle panels). The ligand electron densities are contoured at 1.0σ from an |2Fo| − |Fc| map, and coloured grey (bottom panels). d, Comparison of ligand-binding sites in mGlus and class A GPCRs. The ligands in the crystal structures of mGlu2 TMD–NAM563, mGlu1 TMD–FITM (PDB code 4OR2), mGlu5 TMD–mavoglurant (PDB code 4OO9), β2AR–carazalol (PDB code 2RH1), M2–AF-DX384 (PDB code 5ZKB) and D3R–eticlopride (PDB code 3PBL) are shown as sticks. Only the receptor in the mGlu2 TMD–NAM563 structure is shown in cyan cartoon representation for clarity. e, Comparison of the helical bundles in the inactive and active structures of mGlu2. The helical bundles in the structures of mGlu2 TMD–NAM563, inactive mGlu2, agonist- and PAM-bound mGlu2, and the Gi-free (free) and Gi-bound (G) subunits in the mGlu2–Gi complex (PDB code 7E9G) are shown in an intracellular view. f, Comparison of helix VI in the mGlu2 TMD–NAM563 and mGlu2–Gi structures. The red arrow indicates the downward shift of helix VI in the Gi-bound subunit of the mGlu2–Gi complex relative to the mGlu2 TMD–NAM563 structure and the Gi-free subunit in the Gi-bound structure. g, Comparison of the W6.50 conformation in the inactive and active mGlu2 structures. The mGlu2 TMD–NAM563 structure and the Gi-bound subunit in the mGlu2–Gi structure are coloured cyan and dark red, respectively. The residue W7736.50 in the two structures and the residues that form interactions with W7736.50 in the mGlu2 TMD–NAM563 structure are shown as sticks. The red arrow indicates the rotamer conformational change of W7736.50 in the active structure relative to the inactive structure. h, Comparison of the ionic locks in the inactive and active mGlu2 structures. The mGlu2 TMD–NAM563 structure is coloured cyan. The Gi-bound subunit and Gi protein in the mGlu2–Gi structure are coloured dark red and green, respectively. The residues K6533.50, R6563.53, E754 and E7586.35 that form ionic interactions in the inactive structure are shown as sticks in both structures. The ionic interactions in the inactive structure are shown as red dashed lines and the interaction between R6563.53 and Gαi in the active structure is shown as a blue dashed line.
Extended Data Fig. 7 tr-FRET and functional assays of mGlu2 and mGlu7 homo- and heterodimers.
a, Validation of mGlu2–mGlu7 heterodimer and construct used for structure determination by tr-FRET assay. The signals were measured using the N-terminal HA and Flag tags in each subunit. In each panel, cartoons illustrating the dimer compositions are shown at the bottom. Data are mean ± s.e.m. (bars) from three independent experiments performed in technical triplicate with individual data points shown (dots). b, Cell surface expression of homo- and heterodimers quantified by ELISA. Data were calculated as the fold of mock. Data are mean ± s.e.m. (bars) from three independent experiments performed in technical triplicate with individual data points shown (dots). c–g, Calcium release assays of mGlu2–mGlu7 heterodimer. Data are mean ± s.e.m. from at least three independent experiments performed in technical triplicate and correspond to the peak response. The numbers of independent experiments (n) are shown in parentheses. c, e, f, LY354740-induced calcium release of wild-type mGlu2–mGlu7 heterodimer and mutants. d, g, L-AP4-induced calcium release of wild-type mGlu2–mGlu7 heterodimer and mutants. h, Gi activation of mGlu2–mGlu7(F784S) in the presence of LY354740, LY354740 + mGlu2 PAM (JNJ-40411813) or LY354740 + mGlu7 NAM (MMPIP, ADX71743 or MDIP) measured by the BRET assay. Data are mean ± s.e.m. from at least three independent experiments performed in technical duplicate. The numbers of independent experiments (n) are shown in the parentheses.
Supplementary information
Supplementary Figure
This file contains the raw uncropped gels for Extended Data Figure 1.
Rights and permissions
About this article
Cite this article
Du, J., Wang, D., Fan, H. et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594, 589–593 (2021). https://doi.org/10.1038/s41586-021-03641-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03641-w
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Kinetic fingerprinting of metabotropic glutamate receptors
Communications Biology (2023)
-
Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.