Abstract
Adaptive behaviour necessitates the formation of memories for fearful events, but also that these memories can be extinguished. Effective extinction prevents excessive and persistent reactions to perceived threat, as can occur in anxiety and ‘trauma- and stressor-related’ disorders1. However, although there is evidence that fear learning and extinction are mediated by distinct neural circuits, the nature of the interaction between these circuits remains poorly understood2,3,4,5,6. Here, through a combination of in vivo calcium imaging, functional manipulations, and slice physiology, we show that distinct inhibitory clusters of intercalated neurons (ITCs) in the mouse amygdala exert diametrically opposed roles during the acquisition and retrieval of fear extinction memory. Furthermore, we find that the ITC clusters antagonize one another through mutual synaptic inhibition and differentially access functionally distinct cortical- and midbrain-projecting amygdala output pathways. Our findings show that the balance of activity between ITC clusters represents a unique regulatory motif that orchestrates a distributed neural circuitry, which in turn regulates the switch between high- and low-fear states. These findings suggest that the ITCs have a broader role in a range of amygdala functions and associated brain states that underpins the capacity to adapt to salient environmental demands.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available at: https://data.fmi.ch/PublicationSupplementRepo/. Source data are provided with this paper.
Code availability
Custom-written codes used to analyse data from this study are available upon reasonable request from the corresponding authors.
References
Craske, M. G. et al. Anxiety disorders. Nat. Rev. Dis. Primers 3, 17024 (2017).
Duvarci, S. & Pare, D. Amygdala microcircuits controlling learned fear. Neuron 82, 966–980 (2014).
Milad, M. R. & Quirk, G. J. Fear extinction as a model for translational neuroscience: ten years of progress. Annu. Rev. Psychol. 63, 129–151 (2012).
Orsini, C. A. & Maren, S. Neural and cellular mechanisms of fear and extinction memory formation. Neurosci. Biobehav. Rev. 36, 1773–1802 (2012).
Tovote, P., Fadok, J. P. & Lüthi, A. Neuronal circuits for fear and anxiety. Nat. Rev. Neurosci. 16, 317–331 (2015).
Li, B. Central amygdala cells for learning and expressing aversive emotional memories. Curr. Opin. Behav. Sci. 26, 40–45 (2019).
LeDoux, J. E. Emotion circuits in the brain. Annu. Rev. Neurosci. 23, 155–184 (2000).
Bouton, M. E. Context, ambiguity, and unlearning: sources of relapse after behavioral extinction. Biol. Psychiatry 52, 976–986 (2002).
Herry, C. et al. Neuronal circuits of fear extinction. Eur. J. Neurosci. 31, 599–612 (2010).
Busti, D. et al. Different fear states engage distinct networks within the intercalated cell clusters of the amygdala. J. Neurosci. 31, 5131–5144 (2011).
Collins, D. R. & Paré, D. Spontaneous and evoked activity of intercalated amygdala neurons. Eur. J. Neurosci. 11, 3441–3448 (1999).
Millhouse, O. E. The intercalated cells of the amygdala. J. Comp. Neurol. 247, 246–271 (1986).
Waclaw, R. R., Ehrman, L. A., Pierani, A. & Campbell, K. Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J. Neurosci. 30, 6944–6953 (2010).
Royer, S., Martina, M. & Paré, D. An inhibitory interface gates impulse traffic between the input and output stations of the amygdala. J. Neurosci. 19, 10575–10583 (1999).
Amano, T., Unal, C. T. & Paré, D. Synaptic correlates of fear extinction in the amygdala. Nat. Neurosci. 13, 489–494 (2010).
Likhtik, E., Popa, D., Apergis-Schoute, J., Fidacaro, G. A. & Paré, D. Amygdala intercalated neurons are required for expression of fear extinction. Nature 454, 642–645 (2008).
Asede, D., Bosch, D., Lüthi, A., Ferraguti, F. & Ehrlich, I. Sensory inputs to intercalated cells provide fear-learning modulated inhibition to the basolateral amygdala. Neuron 86, 541–554 (2015).
Mańko, M., Geracitano, R. & Capogna, M. Functional connectivity of the main intercalated nucleus of the mouse amygdala. J. Physiol. (Lond.) 589, 1911–1925 (2011).
Grewe, B. F. et al. Neural ensemble dynamics underlying a long-term associative memory. Nature 543, 670–675 (2017).
Luo, R. et al. A dopaminergic switch for fear to safety transitions. Nat. Commun. 9, 2483 (2018).
Salinas-Hernández, X. I. et al. Dopamine neurons drive fear extinction learning by signaling the omission of expected aversive outcomes. eLife 7, e38818 (2018).
Strobel, C., Marek, R., Gooch, H. M., Sullivan, R. K. P. & Sah, P. Prefrontal and auditory input to intercalated neurons of the amygdala. Cell Rep. 10, 1435–1442 (2015).
Herry, C. et al. Switching on and off fear by distinct neuronal circuits. Nature 454, 600–606 (2008).
Paré, D. & Smith, Y. GABAergic projection from the intercalated cell masses of the amygdala to the basal forebrain in cats. J. Comp. Neurol. 344, 33–49 (1994).
Tovote, P. et al. Midbrain circuits for defensive behaviour. Nature 534, 206–212 (2016).
Senn, V. et al. Long-range connectivity defines behavioral specificity of amygdala neurons. Neuron 81, 428–437 (2014).
McGarry, L. M. & Carter, A. G. Inhibitory gating of basolateral amygdala inputs to the prefrontal cortex. J. Neurosci. 36, 9391–9406 (2016).
Arruda-Carvalho, M. & Clem, R. L. Prefrontal-amygdala fear networks come into focus. Front. Syst. Neurosci. 9, 145 (2015).
Koyama, M. & Pujala, A. Mutual inhibition of lateral inhibition: a network motif for an elementary computation in the brain. Curr. Opin. Neurobiol. 49, 69–74 (2018).
Machens, C. K., Romo, R. & Brody, C. D. Flexible control of mutual inhibition: a neural model of two-interval discrimination. Science 307, 1121–1124 (2005).
Felsenberg, J. et al. Integration of parallel opposing memories underlies memory extinction. Cell 175, 709–722.e15 (2018).
Solomon, R. L. & Corbit, J. D. An opponent-process theory of motivation. I. Temporal dynamics of affect. Psychol. Rev. 81, 119–145 (1974).
Zhang, X., Kim, J. & Tonegawa, S. Amygdala reward neurons form and store fear extinction memory. Neuron 105, 1077–1093.e7 (2020).
Braak, H. & Braak, E. Neuronal types in the basolateral amygdaloid nuclei of man. Brain Res. Bull. 11, 349–365 (1983).
Rousso, D. L. et al. Two pairs of ON and OFF retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell Rep. 15, 1930–1944 (2016).
Guenthner, C. J., Miyamichi, K., Yang, H. H., Heller, H. C. & Luo, L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784 (2013).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Vardy, E. et al. A new DREADD facilitates the multiplexed chemogenetic interrogation of behavior. Neuron 86, 936–946 (2015).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007).
Shemesh, O. A. et al. Temporally precise single-cell-resolution optogenetics. Nat. Neurosci. 20, 1796–1806 (2017).
Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5, e12727 (2016).
Dana, H. et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Claudi, F. et al. Visualizing anatomically registered data with brainrender. eLife 10, e65751 (2020).
Ghosh, K. K. et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011).
Franklin, K. B. J. & Paxinos, G. The Mouse Brain in Stereotaxic Coordinates (Academic, 1997).
Bukalo, O. et al. Prefrontal inputs to the amygdala instruct fear extinction memory formation. Sci. Adv. 1, e1500251 (2015).
Petreanu, L., Huber, D., Sobczyk, A. & Svoboda, K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 10, 663–668 (2007).
Tanaka, Y., Tanaka, Y., Furuta, T., Yanagawa, Y. & Kaneko, T. The effects of cutting solutions on the viability of GABAergic interneurons in cerebral cortical slices of adult mice. J. Neurosci. Methods 171, 118–125 (2008).
Ting, J. T., Daigle, T. L., Chen, Q. & Feng, G. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol. 1183, 221–242 (2014).
Lerner, T. N. et al. Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162, 635–647 (2015).
Gunaydin, L. A. et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014).
Lopes, G. et al. Bonsai: an event-based framework for processing and controlling data streams. Front. Neuroinform. 9, 7 (2015).
An, B. et al. Amount of fear extinction changes its underlying mechanisms. eLife 6, e25224 (2017).
Baek, J. et al. Neural circuits underlying a psychotherapeutic regimen for fear disorders. Nature 566, 339–343 (2019).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Hagihara, K. M., Murakami, T., Yoshida, T., Tagawa, Y. & Ohki, K. Neuronal activity is not required for the initial formation and maturation of visual selectivity. Nat. Neurosci. 18, 1780–1788 (2015).
Kerlin, A. M., Andermann, M. L., Berezovskii, V. K. & Reid, R. C. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron 67, 858–871 (2010).
Gründemann, J. et al. Amygdala ensembles encode behavioral states. Science 364, eaav8736 (2019).
Acknowledgements
We thank F. Ferraguti for discussions on ITC cluster nomenclature; J. Felsenberg, P. Tovote and A. Uematsu for reading and commenting on earlier versions of the manuscript; T. Mrsic-Flogel, M. Häusser and B. Roska for discussion and serving as thesis committee members for K.M.H.; E. Susaki for sharing the CUBIC protocol; Y. Tanaka for sharing the NMDG-based slice preparation protocol; L. Choo for slice physiology demonstration; the GENIE Project for making GCaMP6f, jGCaMP7f and jRGECO1a materials available; B. Roth for making KORD and DREADD materials available; K. Deisseroth for making the ChR2 material available; E. Boyden for making the soCoChR material available; all members of the Lüthi, Holmes, and Ehrlich laboratories for discussion and support, especially T. Eichlisberger for excellent animal care; and the FMI animal facility and microscopy core facility (FAIM), especially L. Gelman and J. Eglinger, and the IT department for constant support. This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 669582), and a Swiss National Science Foundation core grant (310030B_170268) (all to A. Lüthi); by the NIAAA Intramural Research Program (to A.H.); by the German Research Foundation DFG EH 197/3-1 (to I.E.); and by the Novartis Research Foundation.
Author information
Authors and Affiliations
Contributions
K.M.H., O.B., A. Lüthi and A.H. conceived the project. K.M.H. performed all the calcium imaging experiments, virus-based circuit mapping, and the slice experiments in Fig. 4e–j, Extended Data Figs. 11, 12 and analysed the data. O.B. performed chemogenetic manipulation experiments and analysed the data with A. Limoges, T.R., T.C., A.M. and C.W. M.Z. and A.A.-A. performed the slice experiments in Fig. 4a–d, Extended Data Figs. 7, 10 and analysed the data. I.E. conceived the slice experiments and supervised M.Z. and A.A.-A. N.K. performed fibre photometry with K.M.H. M.M. helped to establish the slice physiology setup and provided K.M.H. with technical training and advice on opsin selection. L.S.Z. and R.D.P. provided the Foxp2-Cre mouse line. K.M.H. prepared figures and drafted the manuscript. K.M.H., A. Lüthi and A.H. wrote the manuscript. All the authors commented on and finalized the manuscript and figures. A. Lüthi and A.H. supervised the entire project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Vadim Bolshakov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 3D reconstruction of individual ITC clusters.
a, Schematic of a mouse brain volume. R, rostral; C, caudal; D, dorsal; V, ventral. b, 3D reconstructions were separately obtained via 1) anti-FOXP2 immunostaining; 2) a FoxP2-Cre × Ai14 mouse; and 3) a D1R-Cre × Ai14 mouse, in triplicate for each method. ITCap, apical ITC; ITCl, lateral ITC; ITCav, anteroventral ITC; ITCint, internal ITC; ITCpv, posteroventral ITC; M, medial; L, lateral; Scale bar, 300 μm. c, Example planes of cleared 3D tissue obtained from a wild-type mouse stained with an anti-FOXP2 antibody covering the anterior–posterior axis of ITC clusters. Bregma levels are indicated above each panel. Scale bar, 300 μm. See also Supplementary Video 1 and Supplementary Table 1.
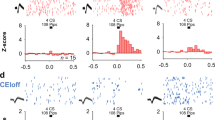
Extended Data Fig. 2 Ca2+ imaging from ITCdm and ITCvm.
a–c, Histological validation of GCaMP6f expression. Wild-type mice (not used for in vivo imaging; n = 2) were injected with AAV-CaMK2-GCaMP6f and killed after 1 month of expression. Thin slices (120 μm) were cut and stained with an anti-FOXP2 antibody. In addition to BLA and CeA neurons, most of the FOXP2-positive ITCdm neurons expressed GCaMP6f. Blue arrow shows a putative large, FOXP2-negative ITC neuron. Scale bars, 500 μm (a), 100 μm (b), and 10 μm (c). d, Example Ca2+ traces from neurons in the CeA, ITCdm, and BLA that were simultaneously imaged with a miniature microscope during fear conditioning. Data obtained from the same mice as in Fig. 1d–g. Grey shading indicates CS presentation (30 s); red line indicates footshock US presentation (1 s). e, f, Histological validation of GCaMP6f expression in ITCvm neurons. Foxp2-Cre mice were injected with an AAV encoding Cre-dependent GCaMP6f, implanted with a GRIN lens, and killed after behavioural experiments. Thin slices (120 μm) were cut and stained with an anti-FOXP2 antibody. Scale bars, 200 μm (e), 50 μm (f). Similar results were obtained with all six mice. g, Summary of histologically confirmed GRIN lens implantation locations for ITCvm recordings. Animals with off-target implantations were excluded from analysis. h, Example Ca2+ traces from neurons in the ITCvm cluster during fear conditioning. Grey shading indicates CS presentation (30 s); red line indicates footshock US presentation (1 s). Images of GCaMP6f expression from the same mouse are shown in e, f. i, j, Correlation matrices of all simultaneously recorded neuron pairs in CeA, ITCdm, and BLA (i), or in ITCvm (j) from representative animals. The entire recording session (11 min) was used. k, Distributions of correlation coefficients from CeA–CeA, ITCdm–ITCdm, and BLA–BLA pairs. Arrowheads indicate medians of the distributions. l, Distribution of correlation coefficients from ITCvm–ITCvm pairs. Arrowhead indicates median of the distribution. m, Summary of medians of correlation coefficient distributions shown in k. Solid lines indicate individual animals in which CeA, the ITCdm cluster, and BLA were simultaneously imaged (n = 3). Dotted lines indicate animals in which only the ITCdm cluster and BLA were simultaneously imaged (n = 5). *P = 0.007, one-way ANOVA followed by Tukey–Kramer test. n, Medians of correlation coefficient distribution. The same analysis as in m was applied to data from home-cage recording sessions. ITCdm neurons also show a trend towards a higher correlation in the absence of CS or US stimulation. *P = 0.12, one-way ANOVA.
Extended Data Fig. 3 Two-photon imaging and fibre photometry.
a, Schematic showing dual-colour in vivo 2-photon imaging through the implanted GRIN lens. ITCdm neurons were labelled by co-injection of an AAV expressing Cre-dependent tdTomato in Foxp2-Cre mice. A GRIN lens was implanted above ITCdm and surrounding BLA and CeA to record Ca2+ responses to aversive skin shocks (USs). b, Mean projected FOV. Green, GCaMP6; magenta, tdTomato. Dashed lines indicate the intermediate capsule surrounding ITCdm. c, Heatplot of Ca2+ responses (ΔF/F) to US presentations showing clustered activation of ITCdm neurons. d, Regions of interest (ROIs) corresponding to ITCdm (magenta) and BLA (blue) neurons. ROI numbers correspond to traces shown in e. e, Example Ca2+ traces from ITCdm and BLA neurons. Red lines indicate US presentations (1 s). We confirmed that face skin shock used in this experiment and footshock similarly activated ITCdm neurons (data not shown). f, Schematic illustrating in vivo fibre photometry in a freely moving mouse. g, ITCdm and ITCvm clusters were targeted with AAVs encoding Cre-dependent jRGECO1a and jGCaMP7f, respectively. Recording fibres were placed in BLA to simultaneously monitor axon terminal Ca2+ dynamics of ITCdm and ITCvm axons. Isosbestic control signals were recorded in the blue channel. h, Example traces of simultaneously recorded dual-colour Ca2+ signals and a control signal during a fear conditioning session. i, Cross-correlation traces between two simultaneously recorded Ca2+ signals. Dark grey lines represent five-trial averaged traces and light grey lines represent individual trials. j, Top, minimum peak values of cross correlation. Bottom, lags of the minimum peak points (n = 2 mice). k, l, Activity heatplot of trial-averaged responses (left, first 5 trials; right, last 5 trials) to footshock US omissions of all the recorded ITCdm (k; n = 271 neurons, from 9 mice) or ITCvm neurons (l; n = 372 neurons, from 6 mice) aligned by CS offset. Cells were sorted according to their averaged ΔF/F responses in the first five trials.
Extended Data Fig. 4 Population activity of ITCdm and ITCvm neurons.
a, b, Heatplots of all recorded neurons: a, 271 ITCdm neurons from 9 mice and b, 372 ITCvm neurons from 6 mice throughout the entire 5-day fear conditioning–extinction paradigm. Neurons were sorted by their classification into fear, extinction, extinction resistant, and non-responsive neurons. c, Example Ca2+ traces from a fear neuron and an extinction neuron from ITCdm and ITCvm, respectively, for all time points indicated in a, b. Habituation, Ext1: 1–5, Ext2: 21–25, and Retrieval time points are duplicated in Fig. 2c. d, Scatter plots visualizing distributions of tone responses of all the recorded ITCdm neurons (left) and ITCvm neurons (middle and right) during Ext1: 1–5 and Ext 2: 21–25 trials. Functional cell-types (fear, extinction, and extinction-resistant neurons) are plotted with different colours; non-responsive neurons in grey (compare with Fig. 2d).
Extended Data Fig. 5 Properties of ITCdm and ITCvm neurons during fear extinction.
a, Trial-averaged ΔF/F Ca2+ responses of ITCdm fear and ITCvm extinction neurons aligned to freezing onset and offset. b, Schematics illustrating the analysis shown c. Correlation coefficients between trial-by-trial (in total, 50 CS trials) freezing levels and CS-response amplitudes of all the recorded neurons across two days of extinction. Distributions of those correlation coefficients (one value from each neuron) were first normalized in each animal, then mean ± s.e.m. values were acquired from all ITCdm and ITCvm recordings and visualized. c, The distribution of correlation coefficients for ITCdm neurons was skewed towards 1, indicating that a larger fraction showed a CS response pattern positively correlated with freezing behaviour. By contrast, ITCvm neurons exhibited the opposite tendency; a response pattern anti-correlated with freezing behaviour. *P = 1.64 × 10−40, Kolmogorov–Smirnov test. d, Trial-averaged ΔF/F Ca2+ responses of ITCvm neurons aligned to tone on habituation (day 1). e, Left, averaged Ca2+ responses to tone CS onset in ITCdm. Vertical dotted lines indicate onsets and 2-s time-window of CSs for AUC analysis. Middle, single-trial average of all the ITCdm neurons. Right, AUC quantification of single-trial responses shown in the middle panel. P = 0.027, one-way ANOVA. f, Trial-averaged ΔF/F Ca2+ responses of ITCdm fear and ITCvm extinction neurons aligned to US omission on day 3. Early, CS1–5; late, CS21–25. Dotted red boxes indicate the expected timing of US delivery. g, Trial-averaged ΔF/F Ca2+ responses of all recorded ITCvm neurons aligned to US omission on day 5. The dotted red box indicates the expected timing of US delivery. h, Relationship between CS responses during last five trials of extinction training on day 4 and US omission responses on day 3 in ITCvm extinction neurons shows a weak positive correlation.
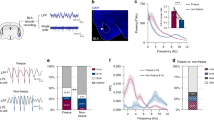
Extended Data Fig. 6 Simultaneous photometry recordings from ITCdm and ITCvm during state transition.
a, Schematic of an elevated zero-maze apparatus. b, Top, example Z-scored Ca2+ traces of simultaneously recorded ITCdm (magenta) and ITCvm (green) neurons with the corresponding location in the maze indicated with grey (closed quadrant) and blue (open quadrant) shading. Bottom, difference between the ITCdm and ITCvm signals. c, Left, percentage of time spent in closed and open quadrants. Right, total number of transitions from closed to open quadrants (15 ± 3.9, mean ± s.e.m.) or from open to closed quadrants. n = 5 mice. Centre line, median; box limits, 25th and 75th percentiles; whiskers, range. d, Averaged activity of ITCdm or ITCvm neurons in the closed or open quadrants did not correlate with the total time spent in open quadrants. Regarding the larger variability in ITC activities in open quadrants, we note that the time an animal spent in open quadrants was, on average, much shorter than that in closed quadrants (c). As such, activity in open quadrants was to a lesser extent averaged out, resulting in higher variability. Eight sessions from n = 5 mice. e, Top, averaged Ca2+ traces during transitions between quadrants. Changes in the balance between ITCdm and ITCvm parallel a transition from a closed to an open quadrant. Bottom, difference between the ITCdm and ITCvm signals. Briefly stated, the results of this experiment make at least three points: 1) the role of the ITC clusters extends beyond signalling acute, cue-triggered fear states to conditioned fear stimuli to encompass state transitions during exploration of a potentially threatening environment; 2) the clusters exhibit markedly divergent, opposing responses to transitions between protected and unprotected environments, as they do in response to conditioned and extinguished fear cues; and 3) the pattern of results shows that increased ITCvm activity occurs when the animal moves from the protected, closed, to the unprotected, open, quadrants. Potentially, this increase in ITCvm neuron activity may serve to inhibit defensive behaviour and thereby enable exploration of the unprotected open quadrants, analogous to the inhibition of freezing behaviour following extinction. Such cross-task neuronal function is not without precedent; for example, BLA neuronal activity during exploration of the open arm in an elevated plus-maze is anti-correlated with conditioned freezing behaviour in the same mice60.
Extended Data Fig. 7 Ex vivo validation of KORD.
a, Fluorescence (mCitrine, left) and infrared (IR, right) images from a slice used for recordings in a Foxp2-Cre mouse injected with AAV-DIO-KORD-mCitrine into ITCdm and ITCvm. Inset, infrared image from a recorded ITCdm neuron. Scale bars, 200 μm. b, Representative current-clamp traces illustrating supra-threshold responses to a +60-pA current injection and continuous recording of the resting membrane potential (RMP) during application of Salvinorin B (SalB, 100 nM) from control (mCitrine−) and KORD-infected (mCitrine+) ITC neurons. Scale bars, RMP, 5 mV, 1 min; current steps, 20 mV, 200 ms. c, SalB application induced significant hyperpolarization of the membrane in mCitrine+ neurons (n = 10 neurons from 4 mice) at both time points versus baseline (12.5 min, *P = 0.003; 17.5 min, *P = 0.0001; two-sided Dunnett’s test). Changes in membrane potential were not significant in the mCitrine− control neurons (n = 5 neurons from 3 mice) at both time points versus baseline (12.5 min, P = 0.99; 17.5 min, P = 0.84; Dunnett’s test). Mean ± s.e.m. d, Comparison of membrane potential changes (ΔRMP) 17.5 min after application of SalB. Changes in KORD-infected neurons (mCitrine+) were significantly different from control (*P = 0.013, Wilcoxon rank-sum test; n = 5 neurons from 3 mice and n = 11 neurons from 5 mice for mCitrine− and mCitrine+, respectively). Mean ± s.e.m. e, Comparisons of spike rates in response to +60-pA current injections before and 17.5 min after application of SalB. KORD-infected neurons (mCitrine+, n = 11) significantly reduced spike rate (from 26.2 ± 1.69 Hz to 20.1 ± 3.30 Hz; *P = 0.016, Wilcoxon signed-rank test). By contrast, mCitrine− neurons (n = 4 neurons from 3 mice) did not show a significant reduction in spike rate (from 25.3 ± 2.89 Hz to 23.7 ± 2.27 Hz; P = 0.25, Wilcoxon signed-rank test. Mean ± s.e.m.
Extended Data Fig. 8 Histology for chemogenetic manipulations of distinct ITC clusters.
a, Examples of histological validation of KORD–mCitrine expression in ITCvm neurons across multiple anterior-to-posterior coronal sections. Following all the behavioural experiments (Fig. 3), mice were killed and slices (50 μm) cut and stained with an anti-FOXP2 antibody. Scale bar, 200 μm. b, Examples of histological validation of hM3Dq–mCherry expression in ITCvm neurons across multiple anterior-to-posterior coronal sections. Scale bar, 200 μm. c, Heatmaps illustrating virus expression aggregated across Cre+ mice in a and b. Scale bar indicates the fraction of animals exhibiting expression at each locus (0: no mice expressed; 1: all mice expressed). d, Examples of histological validation of KORD–mCitrine expression in ITCdm neurons across multiple anterior-to-posterior coronal sections. Scale bar, 200 μm. e, Examples of histological validation of hM3Dq–mCherry expression in ITCdm neurons across multiple anterior-to-posterior coronal sections. Scale bar, 200 μm. f, Heatmaps illustrating virus expression aggregated across Cre+ mice in d and e. Scale bar indicates the fraction of animals exhibiting expression at each locus (0: no mice expressed; 1: all mice expressed). Bregma levels are noted above the panels in a and d.
Extended Data Fig. 9 Additional chemogenetic manipulations of ITCdm and ITCvm clusters.
a, Chemogenetic manipulations. AAV encoding Cre-dependent hM3Dq was targeted to ITCdm neurons in Foxp2-Cre mice. b, ITCdm activation. Freezing on retrieval: Cre+, 45.7 ± 7.6%; Cre−, 28.5 ± 3.3%; *P = 0.0044. c, Schematic showing the dual-cluster chemogenetic manipulation experiment. An AAV encoding Cre-dependent KORD was targeted to both ITCvm and ITCdm neurons in Foxp2-Cre mice. d, Freezing behaviour of experimental (Cre+) and control (Cre−) mice; controls were injected with the AAV and administered the ligand. Arrows and colour-shadings indicate the timing of SalB administration before extinction retrieval. Freezing levels on extinction retrieval (day 3): Cre+, 46.4 ± 4.3%; Cre−, 27.3 ± 2.7%; *P < 0.0001, repeated-measures ANOVA followed by Sidak post hoc test. N.S., not significantly different on day 1 or day 2 freezing. Mean ± s.e.m. e, Examples of histological validation of KORD–mCitrine expression in ITCvm and ITCdm neurons across multiple anterior-to-posterior coronal sections. Following behavioural experiments, mice were killed and slices (50 μm) cut and stained with an anti-FOXP2 antibody. Scale bar, 200 μm. f, Heatmaps illustrating virus expression aggregated across Cre+ mice. Scale bar indicates the fraction of animals exhibiting expression at each locus (0: no mice expressed; 1: all mice expressed).
Extended Data Fig. 10 Reciprocal inhibitory connections between ITC clusters.
a, Inter-cluster virus-based tracing (ITCdm-targeted example shown, same strategy for ITCvm targeting). AAV encoding Cre-dependent synaptophysin–GFP targeted to ITCdm or ITCvm neurons in Foxp2-Cre mice. b, c, Confocal images from ITCdm (b) or ITCvm (c) injections. FOXP2 expression visualized using immunohistochemistry (IHC). Injections repeated in five (ITCdm), and four (ITCvm) mice. Scale bars: 500 μm (b1, c1); 200 μm (b2, 3, c2, 3). d, e, Histological validation of selective ChR2 expression in ITCdm (d1–3) and ITCvm (e1–3). Scale bars: 500 μm (d1, e1); 50 μm (d3, e3). Repeated for all mice shown in Fig. 4. f, Representative example of IPSCs recorded at different holding potentials from an ITCdm neuron upon photostimulation of ChR2-EYFP fibers from ITCvm. g, i, Schematics illustrating ex vivo slice electrophysiological experiments with direct cluster stimulation. The ITCvm (g) or ITCdm (i) cluster was targeted with an AAV encoding Cre-dependent ChR2. Recordings were performed in the other cluster while the ChR2-positive cluster or the surrounding area was selectively stimulated with aperture-restricted stimulation (see Methods). h, j, Images taken from the recording setup illustrating positions of on- and off-cluster stimulation for ITCvm while recording in ITCdm and vice versa. k, Summary of PTX application experiments (n = 3 from 3 mice). Magenta lines represent ITCvm→ITCdm connections and green lines represent ITCdm→ITCvm connections. *P = 0.005, paired t-test. l, Summary of experiments shown in g–j. On-cluster photostimulation evoked significantly larger IPSCs than off-cluster stimulation. Note that we used the highest value out of the four off-cluster stimulation sites. *P = 0.0016, paired t-test.
Extended Data Fig. 11 Selective ex vivo ITC cluster stimulation.
a, Histological validation of selective soCoChR expression. Scale bars, 50 μm. Repeated for n = 3 mice. b, Schematic illustrating ex vivo slice electrophysiological experiments. ITCdm and ITCvm neurons were targeted with an AAV encoding Cre-dependent soCoChR. Two optical fibres for photostimulation were placed above the ITC clusters. Whole-cell recordings were performed from neurons in the ITCdm, ITCvm, and ITCl clusters. To block synaptic currents, CPP (10 μM), CNQX (10 μM) and PTX (100 μM) were applied. c–e, Example responses to photostimulation of ITCdm or ITCvm clusters recorded under current-clamp configuration from ITCdm (c), ITCvm (d), and ITCl (e) neurons. f, Summary of experiments shown in b–e. All recorded ITCdm and ITCvm neurons fired action potentials selectively in response to photostimulation with the corresponding fibre. ITCl neurons did not fire action potentials upon photostimulation of ITCdm or ITCvm clusters. g, Schematic illustrating experiments wherein the position of the optical fibre was systematically moved away from the optimal location (X = 0 μm). To block synaptic currents, CPP (10 μM), CNQX (10 μM), and PTX (100 μM) were applied. These experiments further confirmed the spatial resolution of the photostimulation configuration. h, Example voltage-clamp recordings from an ITC neuron showing IPSCs evoked at different positions of the optical fibre. i, Left, quantification of peak IPSC amplitudes of the neuron shown in h. Right, normalized by the value at X = 0 μm. j, Summary of all recorded neurons (n = 3 from 3 mice). k, Summary of additionally performed current-clamp recordings (n = 3 from 3 mice).
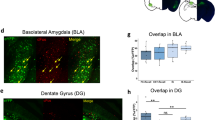
Extended Data Fig. 12 ITC to CeM connectivity.
a, Schematic illustrating the experimental design used to assess connectivity between ITCdm or ITCvm neurons and CeM neurons. To isolate IPSCs, CPP (10 μM) and CNQX (10 μM) were applied. b, Example voltage-clamp recording from a CeM neuron showing selective IPSC elicited by stimulation of the ITCvm but not ITCdm. c, Summary of connectivity from ITCdm and ITCvm to CeM (n = 11 neurons from 4 mice). *P = 7.5 × 10−3, Fisher’s exact test. d, Evoked IPSC amplitudes recorded from CeM neurons (at −70 mV holding potential) in response to stimulation of ITCdm or ITCvm clusters. e, Synaptic conductance calculated based on IPSC amplitudes recorded at three different holding potentials (−60, −70, and −80 mV). f, Example injections of CTB555 targeted to the ventrolateral periaqueductal grey (vlPAG). Scale bar, 200 μm. g, Schematic illustrating the experimental design used to assess connectivity between ITCdm or ITCvm neurons and CeM neurons projecting to the vlPAG. vlPAG-projecting CeM neurons were retrogradely labelled with CTB injections into the vlPAG. Whole-cell recordings were performed in CTB+ CeM neurons while photostimulating incoming axons expressing ChR2–EYFP from either ITCdm or ITCvm. h, Summary of connectivity from ITCdm and ITCvm to vlPAG-projecting CeM neurons (n = 15 neurons from 4 mice and n = 10 neurons from 4 mice). *P = 0.0001, Fisher’s exact test. i, Evoked IPSC amplitudes recorded from CTB+ CeM neurons (at 0 mV holding potential) in response to stimulation of axons from ITCdm or ITCvm. j, Example injections of CTB555 and CTB647 targeted to the PL or IL, respectively. Scale bar, 500 μm. k, Example BLA histology showing PLp and ILp BLA neurons. Both subpopulations are located in the medial part of the BLA. Scale bar, 200 μm. l, Mostly non-overlapping PLp and ILp subpopulations; orange arrow indicates an example of a double-labelled neuron. Scale bar, 50 μm. m, Quantification of double-labelled neurons (n = 3 mice). Mean ± s.e.m. n, State-dependent functional changes in amygdala circuitry regulated by ITC clusters. Node size indicates activity level, connection width represents putative functional connection strength.
Supplementary information
Supplementary Tables
This file contains Supplementary Table 1, a cross-study comparison of ITC cluster nomenclature and Supplementary Table 2, statistical analyses summary for chemogenetic experiments.
Video 1
: 3D-reconstruction of individual ITC clusters. An example 3D-reconstruction of ITC clusters from a cleared FoxP2-Cre crossed with tdTomato reporter mouse brain. The same sample as shown in Extended Data Fig.1b is presented. Note, this intercross mouse line shows tdTomato signal in some CeA neurons and blood vessels in addition to ITCs, likely because of developmental transient expression of FoxP2.
Video 2
: Post-hoc identification of ITCdm in a 3D confocal image of a cleared tissue. An example confocal 3D image stack of cleared and stained tissue acquired with a 20x objective. The same sample as shown in Fig.1d, e is presented.
Video 3
: Spatially clustered robust aversive shock responses in ITCdm Example Ca2+ movies from simultaneously recorded BLA, CeA, and ITCdm regions. ITCdm shows spatially clustered and robust responses to 5 trials of aversive footshock presentation. The video was sped up 2x. The timing of shock presentation is indicated with red ‘shock’ text. The same sample as shown in Fig.1f, g is presented.
Source data
Rights and permissions
About this article
Cite this article
Hagihara, K.M., Bukalo, O., Zeller, M. et al. Intercalated amygdala clusters orchestrate a switch in fear state. Nature 594, 403–407 (2021). https://doi.org/10.1038/s41586-021-03593-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03593-1
This article is cited by
-
The amygdala NT3-TrkC pathway underlies inter-individual differences in fear extinction and related synaptic plasticity
Molecular Psychiatry (2024)
-
A distinct cortical code for socially learned threat
Nature (2024)
-
Neuronal types in the mouse amygdala and their transcriptional response to fear conditioning
Nature Neuroscience (2023)
-
Optogenetic stimulation of mouse Hoxb8 microglia in specific regions of the brain induces anxiety, grooming, or both
Molecular Psychiatry (2023)
-
The cerebellum regulates fear extinction through thalamo-prefrontal cortex interactions in male mice
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.