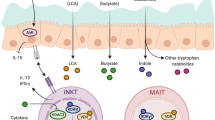
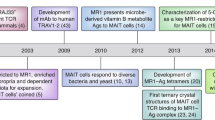
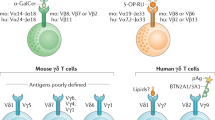
Abstract
The unconventional T cell compartment encompasses a variety of cell subsets that straddle the line between innate and adaptive immunity, often reside at mucosal surfaces and can recognize a wide range of non-polymorphic ligands. Recent advances have highlighted the role of unconventional T cells in tissue homeostasis and disease. In this Review, we recast unconventional T cell subsets according to the class of ligand that they recognize; their expression of semi-invariant or diverse T cell receptors; the structural features that underlie ligand recognition; their acquisition of effector functions in the thymus or periphery; and their distinct functional properties. Unconventional T cells follow specific selection rules and are poised to recognize self or evolutionarily conserved microbial antigens. We discuss these features from an evolutionary perspective to provide insights into the development and function of unconventional T cells. Finally, we elaborate on the functional redundancy of unconventional T cells and their relationship to subsets of innate and adaptive lymphoid cells, and propose that the unconventional T cell compartment has a critical role in our survival by expanding and complementing the role of the conventional T cell compartment in protective immunity, tissue healing and barrier function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Godfrey, D. I., MacDonald, H. R., Kronenberg, M., Smyth, M. J. & Van Kaer, L. NKT cells: what’s in a name? Nat. Rev. Immunol. 4, 231–237 (2004).
Lu, L., Werneck, M. B. F. & Cantor, H. The immunoregulatory effects of Qa-1. Immunol. Rev. 212, 51–59 (2006).
Colmone, A. & Wang, C.-R. H2-M3-restricted T cell response to infection. Microbes Infect. 8, 2277–2283 (2006).
Bendelac, A., Savage, P. B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Cohen, N. R., Garg, S. & Brenner, M. B. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv. Immunol. 102, 1–94 (2009).
Cheroutre, H., Lambolez, F. & Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456 (2011).
Kim, H.-J. & Cantor, H. Regulation of self-tolerance by Qa-1-restricted CD8+ regulatory T cells. Semin. Immunol. 23, 446–452 (2011).
Godfrey, D. I., Uldrich, A. P., McCluskey, J., Rossjohn, J. & Moody, D. B. The burgeoning family of unconventional T cells. Nat. Immunol. 16, 1114–1123 (2015).
Mayassi, T. & Jabri, B. Human intraepithelial lymphocytes. Mucosal Immunol. 11, 1281–1289 (2018).
Ogg, G., Cerundolo, V. & McMichael, A. J. Capturing the antigen landscape: HLA-E, CD1 and MR1. Curr. Opin. Immunol. 59, 121–129 (2019).
Legoux, F., Salou, M. & Lantz, O. MAIT cell development and functions: the microbial connection. Immunity 53, 710–723 (2020).
Vantourout, P. & Hayday, A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13, 88–100 (2013).
Nielsen, M. M., Witherden, D. A. & Havran, W. L. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol. 17, 733–745 (2017).
Jouand, N. et al. HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLoS Pathog. 14, e1007041 (2018).
Bian, Y. et al. MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLoS Pathog. 13, e1006384 (2017).
Bendelac, A. et al. CD1 recognition by mouse NK1+ T lymphocytes. Science 268, 863–865 (1995). This paper demonstrates that NKT cells recognize CD1d.
Mattner, J. et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434, 525–529 (2005). This paper and the following reference show that NKT cells promote defence against bacterial infections by recognizing either self or microbial glycolipids.
Kinjo, Y. et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434, 520–525 (2005).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). This paper demonstrates that MR1 presents vitamin B metabolites to MAIT cells.
Corbett, A. J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014).
Shawar, S. M., Rodgers, J. R., Cook, R. G. & Rich, R. R. Specialized function of the nonclassical MHC class I molecule Hmt: a specific receptor for N-formylated peptides. Immunol. Res. 10, 365–375 (1991).
Lenz, L. L., Dere, B. & Bevan, M. J. Identification of an H2-M3-restricted Listeria epitope: implications for antigen presentation by M3. Immunity 5, 63–72 (1996). This paper shows that the non-classical MHC molecule H2-M3 presents bacterial formylated peptides.
Tanaka, Y. et al. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature 375, 155–158 (1995).
de Jong, A. et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol. 15, 177–185 (2014).
Birkinshaw, R. W. et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol. 16, 258–266 (2015). The crystal structures in this paper show the breaking of the co-recognition paradigm as defined by the requirement for a TCR to simultaneously recognize an antigen and the antigen-presenting molecule.
Dimova, T. et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc. Natl Acad. Sci. USA 112, E556–E565 (2015).
Tomasec, P. et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000).
Braud, V., Jones, E. Y. & McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 27, 1164–1169 (1997).
Jabri, B. et al. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity 17, 487–499 (2002).
Braud, V. M. et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998).
Oliveira, C. C. et al. The nonpolymorphic MHC Qa-1b mediates CD8+ T cell surveillance of antigen-processing defects. J. Exp. Med. 207, 207–221 (2010).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Barbee, S. D. et al. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc. Natl Acad. Sci. USA 108, 3330–3335 (2011). This paper identifies SKINT-1 as the selecting ligand for DETCs.
Di Marco Barros, R. et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ t cell compartments. Cell 167, 203–218 (2016). This paper identifies BTNL molecules as the selecting ligands for gut-specific γδ T cells.
Mayassi, T. et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell 176, 967–981 (2019). This paper characterizes the functional and transcriptional program of the gut innate-like human BTNL-specific γδ T cells and shows how chronic inflammation permanently reshapes these niches.
Groh, V., Steinle, A., Bauer, S. & Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science 279, 1737–1740 (1998).
Kong, Y. et al. The NKG2D ligand ULBP4 binds to TCRγ9/δ2 and induces cytotoxicity to tumor cells through both TCRγδ and NKG2D. Blood 114, 310–317 (2009).
Crowley, M. P., Reich, Z., Mavaddat, N., Altman, J. D. & Chien, Y. The recognition of the nonclassical major histocompatibility complex (MHC) class I molecule, T10, by the γδ T cell, G8. J. Exp. Med. 185, 1223–1230 (1997).
Shin, S. et al. Antigen recognition determinants of γδ T cell receptors. Science 308, 252–255 (2005).
Lantz, O. & Bendelac, A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med. 180, 1097–1106 (1994).
Kawano, T. et al. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science 278, 1626–1629 (1997).
Tilloy, F. et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 (1999).
Porcelli, S., Yockey, C. E., Brenner, M. B. & Balk, S. P. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8– α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J. Exp. Med. 178, 1–16 (1993).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003).
Van Rhijn, I. et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol. 14, 706–713 (2013).
Gras, S. et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat. Commun. 7, 13257 (2016).
Melandri, D. et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 19, 1352–1365 (2018).
Castro, C. D., Luoma, A. M. & Adams, E. J. Coevolution of T-cell receptors with MHC and non-MHC ligands. Immunol. Rev. 267, 30–55 (2015).
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796 (2018). This paper demonstrates the role of H2-M3-restricted T cell responses in regulating the microbiota and wound healing in the skin.
Grant, E. J. et al. The unconventional role of HLA-E: the road less traveled. Mol. Immunol. 120, 101–112 (2020).
McDonald, B. D., Bunker, J. J., Ishizuka, I. E., Jabri, B. & Bendelac, A. Elevated T cell receptor signaling identifies a thymic precursor to the TCRαβ+CD4−CD8β− intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014).
Mayans, S. et al. αβT cell receptors expressed by CD4−CD8αβ− intraepithelial T cells drive their fate into a unique lineage with unusual MHC reactivities. Immunity 41, 207–218 (2014).
Park, S. H. et al. Selection and expansion of CD8α/α1 T cell receptor α/β1 intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J. Exp. Med. 190, 885–890 (1999).
Gherardin, N. A., McCluskey, J., Rossjohn, J. & Godfrey, D. I. The diverse family of MR1-restricted T cells. J. Immunol. 201, 2862–2871 (2018).
Uldrich, A. P. et al. CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol. 14, 1137–1145 (2013).
Luoma, A. M. et al. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity 39, 1032–1042 (2013).
Le Nours, J. et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366, 1522–1527 (2019).
Garcia, K. C. et al. An αβ T cell receptor structure at 2.5 A and its orientation in the TCR–MHC complex. Science 274, 209–219 (1996).
Garboczi, D. N. et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Beringer, D. X. et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 16, 1153–1161 (2015).
Gras, S. et al. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity 45, 749–760 (2016).
La Gruta, N. L., Gras, S., Daley, S. R., Thomas, P. G. & Rossjohn, J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 18, 467–478 (2018).
Adams, E. J. & Luoma, A. M. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31, 529–561 (2013).
Van Rhijn, I., Godfrey, D. I., Rossjohn, J. & Moody, D. B. Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 15, 643–654 (2015).
Saunders, P. M. et al. A bird’s eye view of NK cell receptor interactions with their MHC class I ligands. Immunol. Rev. 267, 148–166 (2015).
Hoare, H. L. et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7, 256–264 (2006).
Borg, N. A. et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007). This paper provides the first insights into how unconventional TCR recognition differs from that of conventional TCRs in the form of type I NKT TCR–CD1d–lipid complex.
Rossjohn, J., Pellicci, D. G., Patel, O., Gapin, L. & Godfrey, D. I. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12, 845–857 (2012).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
Eckle, S. B. G. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211, 1585–1600 (2014).
Patel, O. et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat. Immunol. 13, 857–863 (2012).
Girardi, E. et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat. Immunol. 13, 851–856 (2012).
Gherardin, N. A. et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44, 32–45 (2016).
Wun, K. S. et al. T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat. Immunol. 19, 397–406 (2018).
Bendelac, A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182, 2091–2096 (1995).
Seach, N. et al. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol. 191, 6002–6009 (2013).
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). This paper demonstrates the critical role of PLZF in the effector programming of unconventional T cells such as NKT cells and MAIT cells.
Koay, H.-F. et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 4, eaay6039 (2019).
Griewank, K. et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27, 751–762 (2007).
Urdahl, K. B., Sun, J. C. & Bevan, M. J. Positive selection of MHC class Ib-restricted CD8+ T cells on hematopoietic cells. Nat. Immunol. 3, 772–779 (2002).
Bediako, Y. et al. SAP is required for the development of innate phenotype in H2-M3-restricted CD8+ T cells. J. Immunol. 189, 4787–4796 (2012).
McDonald, B. D., Jabri, B. & Bendelac, A. Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 18, 514–525 (2018).
Wei, D. G. et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J. Exp. Med. 202, 239–248 (2005).
Guy-Grand, D. et al. Origin, trafficking, and intraepithelial fate of gut-tropic T cells. J. Exp. Med. 210, 1839–1854 (2013).
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). This paper demonstrates that there is a window in early life for the colonization of tissues by MAIT cells and stresses the role of MAIT cells in wound repair.
Mazzarino, P. et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur. J. Immunol. 35, 3240–3247 (2005).
Doorduijn, E. M. et al. T cells engaging the conserved MHC class Ib molecule Qa-1b with TAP-independent peptides are semi-invariant lymphocytes. Front. Immunol. 9, 60 (2018).
Havran, W. L. & Allison, J. P. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature 344, 68–70 (1990).
Benlagha, K., Weiss, A., Beavis, A., Teyton, L. & Bendelac, A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191, 1895–1904 (2000).
Dusseaux, M. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011).
Mueller, S. N. & Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16, 79–89 (2016).
Masopust, D. & Soerens, A. G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 37, 521–546 (2019).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012).
Chan, A. C. et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4+ and CD4− subsets. Clin. Exp. Immunol. 172, 129–137 (2013).
Loh, L. et al. Human mucosal-associated invariant T cells in older individuals display expanded TCRαβ clonotypes with potent antimicrobial responses. J. Immunol. 1950, 1119–1133 (2020).
Zaid, A. et al. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl Acad. Sci. USA 111, 5307–5312 (2014).
Hayday, A., Theodoridis, E., Ramsburg, E. & Shires, J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat. Immunol. 2, 997–1003 (2001).
Hinks, T. S. C. et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28, 3249–3262.e5 (2019).
Legoux, F., Salou, M. & Lantz, O. Unconventional or preset αβ T cells: evolutionarily conserved tissue-resident T cells recognizing nonpeptidic ligands. Annu. Rev. Cell Dev. Biol. 33, 511–535 (2017).
Guy-Grand, D., Cuénod-Jabri, B., Malassis-Seris, M., Selz, F. & Vassalli, P. Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur. J. Immunol. 26, 2248–2256 (1996). This paper provides the first evidence that intestinal tissue-resident unconventional intraepithelial T lymphocytes express NK receptors and mediate NK-cell-like killing.
Toulon, A. et al. A role for human skin-resident T cells in wound healing. J. Exp. Med. 206, 743–750 (2009).
Groh, V. et al. Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc. Natl Acad. Sci. USA 96, 6879–6884 (1999).
Pamer, E. G., Bevan, M. J. & Lindahl, K. F. Do nonclassical, class Ib MHC molecules present bacterial antigens to T cells? Trends Microbiol. 1, 35–38 (1993).
Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010).
Gaya, M. et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell 172, 517–533 (2018).
Boismenu, R. & Havran, W. L. Modulation of epithelial cell growth by intraepithelial γδ T cells. Science 266, 1253–1255 (1994). This paper was the first study to propose that TCRγδ T cells have a role in wound healing.
Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 15, 771–783 (2015).
Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J. & Ploegh, H. L. Viral subversion of the immune system. Annu. Rev. Immunol. 18, 861–926 (2000).
Turtle, C. J. et al. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161hi CD8α+ semi-invariant T cells. Blood 118, 2752–2762 (2011).
Slichter, C. K. et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight 1, e86292 (2016).
Wesley, J. D., Tessmer, M. S., Chaukos, D. & Brossay, L. NK cell-like behavior of Vα14i NK T cells during MCMV infection. PLoS Pathog. 4, e1000106 (2008).
Tyznik, A. J. et al. Cutting edge: the mechanism of invariant NKT cell responses to viral danger signals. J. Immunol. 181, 4452–4456 (2008).
Strid, J. et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat. Immunol. 9, 146–154 (2008).
Strid, J., Sobolev, O., Zafirova, B., Polic, B. & Hayday, A. The intraepithelial T cell response to NKG2D-ligands links lymphoid stress surveillance to atopy. Science 334, 1293–1297 (2011).
Wu, Y. et al. An innate-like Vδ1+ γδ T cell compartment in the human breast is associated with remission in triple-negative breast cancer. Sci. Transl. Med. 11, eaax9364 (2019).
Carnaud, C. et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J. Immunol. 163, 4647–4650 (1999).
Godfrey, D. I., Le Nours, J., Andrews, D. M., Uldrich, A. P. & Rossjohn, J. Unconventional T cell targets for cancer immunotherapy. Immunity 48, 453–473 (2018).
Crowther, M. D. et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 21, 178–185 (2020).
Edholm, E.-S., Banach, M. & Robert, J. Evolution of innate-like T cells and their selection by MHC class I-like molecules. Immunogenetics 68, 525–536 (2016).
Mondot, S., Boudinot, P. & Lantz, O. MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics 68, 537–548 (2016).
Boudinot, P. et al. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl Acad. Sci. USA 113, E2983–E2992 (2016).
Flajnik, M. F. & Kasahara, M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 47–59 (2010).
Rodgers, J. R. & Cook, R. G. MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol. 5, 459–471 (2005).
Van Rhijn, I., Ly, D. & Moody, D. B. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv. Exp. Med. Biol. 783, 181–197 (2013).
van Wilgenburg, B. et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat. Commun. 9, 4706 (2018).
Legoux, F. et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019).
Fischer, A. Severe combined immunodeficiencies. Immunodefic. Rev. 3, 83–100 (1992).
Barreiro, L. B. et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5, e1000562 (2009).
Barreiro, L. B. & Quintana-Murci, L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30 (2010).
Howson, L. J. et al. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci. Immunol. 5, eabc9492 (2020).
Harsha Krovi, S. et al. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun. 11, 6238 (2020).
Constantinides, M. G., McDonald, B. D., Verhoef, P. A. & Bendelac, A. A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014).
Heinzel, A. S. et al. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196, 1473–1481 (2002).
Kabelitz, D. et al. The primary response of human γ/δ+ T cells to Mycobacterium tuberculosis is restricted to Vγ9-bearing cells. J. Exp. Med. 173, 1331–1338 (1991).
Gold, M. C. et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8, e1000407 (2010).
Nish, S. & Medzhitov, R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity 34, 629–636 (2011).
Borbulevych, O. Y., Santhanagopolan, S. M., Hossain, M. & Baker, B. M. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J. Immunol. 187, 2453–2463 (2011).
Wun, K. S. et al. Human and mouse type I natural killer T cell antigen receptors exhibit different fine specificities for CD1d-antigen complex. J. Biol. Chem. 287, 39139–39148 (2012).
Acknowledgements
We thank D. Littler for generating Fig. 1; A. Bendelac for sharing his insights on innate-like lymphocytes; D. Guy-Grand for sharing her insights on intraepithelial lymphocytes over many years; and M. Kronenberg and K. Sangani for discussions. This Review was supported by grants to B.J. from the National Institutes of Health (NIH: R01 DK67180 and R01 DK098435) and the Digestive Diseases Research Core Center at the University of Chicago (P30 DK42086); to J.R. from the Australian ARC Laureate Fellowship; and to L.B.B. from NIH: R01 GM134376.
Author information
Authors and Affiliations
Contributions
B.J. conceived the writing of the article. T.M. and B.J. conceptualized the framework and wrote the article, with input from J.R. on the ‘Modes of ligand recognition’ section and input from L.B.B. on the ‘Evolution and redundancy’ section. All authors reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Laurent Gapin, Paul Klenerman and Laura Mackay for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mayassi, T., Barreiro, L.B., Rossjohn, J. et al. A multilayered immune system through the lens of unconventional T cells. Nature 595, 501–510 (2021). https://doi.org/10.1038/s41586-021-03578-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03578-0
This article is cited by
-
The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes
Nature Immunology (2024)
-
Landscape of unconventional γδ T cell subsets in cancer
Molecular Biology Reports (2024)
-
Unconventional haircare
Nature Immunology (2023)
-
Innate lymphoid cells and innate-like T cells in cancer — at the crossroads of innate and adaptive immunity
Nature Reviews Cancer (2023)
-
Unconventional immune cells in the gut mucosal barrier: regulation by symbiotic microbiota
Experimental & Molecular Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.