Abstract
Vascular malformations are thought to be monogenic disorders that result in dysregulated growth of blood vessels. In the brain, cerebral cavernous malformations (CCMs) arise owing to inactivation of the endothelial CCM protein complex, which is required to dampen the activity of the kinase MEKK31,2,3,4. Environmental factors can explain differences in the natural history of CCMs between individuals5, but why single CCMs often exhibit sudden, rapid growth, culminating in strokes or seizures, is unknown. Here we show that growth of CCMs requires increased signalling through the phosphatidylinositol-3-kinase (PI3K)–mTOR pathway as well as loss of function of the CCM complex. We identify somatic gain-of-function mutations in PIK3CA and loss-of-function mutations in the CCM complex in the same cells in a majority of human CCMs. Using mouse models, we show that growth of CCMs requires both PI3K gain of function and CCM loss of function in endothelial cells, and that both CCM loss of function and increased expression of the transcription factor KLF4 (a downstream effector of MEKK3) augment mTOR signalling in endothelial cells. Consistent with these findings, the mTORC1 inhibitor rapamycin effectively blocks the formation of CCMs in mouse models. We establish a three-hit mechanism analogous to cancer, in which aggressive vascular malformations arise through the loss of vascular ‘suppressor genes’ that constrain vessel growth and gain of a vascular ‘oncogene’ that stimulates excess vessel growth. These findings suggest that aggressive CCMs could be treated using clinically approved mTORC1 inhibitors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. DNA sequencing data are available on request from D.A.M. The data are not publicly available as they contain information that could compromise the privacy and consent of research participants. Public datasets used here are available at COSMIC (cancer.sanger.ac.uk/cosmic), dbSNP (ncbi.nlm.nih.gov/snp), 1000 Genomes Project (internationalgenome.org) and ExAC (gnomad.broadinstitute.org). Source data are provided with this paper.
Code availability
Variant-calling software was implemented as part of Gonomics, an ongoing effort to develop an open-source genomics platform in the Go programming language. Gonomics can be accessed at github.com/vertgenlab/gonomics.
References
Cuttano, R. et al. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 8, 6–24 (2015).
Zhou, Z. et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature 532, 122–126 (2016).
Renz, M. et al. Regulation of β1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev. Cell 32, 181–190 (2015).
Otten, C. et al. Systematic pharmacological screens uncover novel pathways involved in cerebral cavernous malformations. EMBO Mol. Med. 10, e9155 (2018).
Tang, A. T. et al. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature 545, 305–310 (2017).
Fischer, A., Zalvide, J., Faurobert, E., Albiges-Rizo, C. & Tournier-Lasserve, E. Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol. Med. 19, 302–308 (2013).
Fisher, O. S. & Boggon, T. J. Signaling pathways and the cerebral cavernous malformations proteins: lessons from structural biology. Cell. Mol. Life Sci. 71, 1881–1892 (2014).
Plummer, N. W., Zawistowski, J. S. & Marchuk, D. A. Genetics of cerebral cavernous malformations. Curr. Neurol. Neurosci. Rep. 5, 391–396 (2005).
Al-Shahi Salman, R. et al. Untreated clinical course of cerebral cavernous malformations: a prospective, population-based cohort study. Lancet Neurol. 11, 217–224 (2012).
Awad, I. A. & Polster, S. P. Cavernous angiomas: deconstructing a neurosurgical disease. J. Neurosurg. 131, 1–13 (2019).
Porter, P. J., Willinsky, R. A., Harper, W. & Wallace, M. C. Cerebral cavernous malformations: natural history and prognosis after clinical deterioration with or without hemorrhage. J. Neurosurg. 87, 190–197 (1997).
Boulday, G. et al. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J. Exp. Med. 208, 1835–1847 (2011).
Maddaluno, L. et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 498, 492–496 (2013).
Detter, M. R., Snellings, D. A. & Marchuk, D. A. Cerebral cavernous malformations develop through clonal expansion of mutant endothelial cells. Circ. Res. 123, 1143–1151 (2018).
Malinverno, M. et al. Endothelial cell clonal expansion in the development of cerebral cavernous malformations. Nat. Commun. 10, 2761 (2019).
Ramírez-Zamora, A. & Biller, J. Brainstem cavernous malformations: a review with two case reports. Arq. Neuropsiquiatr. 67 (3B), 917–921 (2009).
Castro, M. et al. CDC42 deletion elicits cerebral vascular malformations via increased MEKK3-dependent KLF4 expression. Circ. Res. 124, 1240–1252 (2019).
Hong, C. C. et al. Cerebral cavernous malformations are driven by ADAMTS5 proteolysis of versican. J. Exp. Med. 217, e20200140 (2020).
Lissbrant, I. F., Lissbrant, E., Persson, A., Damber, J. E. & Bergh, A. Endothelial cell proliferation in male reproductive organs of adult rat is high and regulated by testicular factors. Biol. Reprod. 68, 1107–1111 (2003).
Samuels, Y. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573 (2005).
Rodriguez-Laguna, L. et al. Somatic activating mutations in PIK3CA cause generalized lymphatic anomaly. J. Exp. Med. 216, 407–418 (2019).
Castillo, S. D. et al. Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med. 8, 332ra43 (2016).
Castel, P. et al. Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med. 8, 332ra42 (2016).
Luks, V. L. et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 166, 1048–1054 (2015).
Limaye, N. et al. Somatic activating PIK3CA mutations cuse venous malformation. Am. J. Hum. Genet. 97, 914–921 (2015).
Storck, S. E. et al. Endothelial LRP1 transports amyloid-β(1-42) across the blood-brain barrier. J. Clin. Invest. 126, 123–136 (2016).
Rigamonti, D. et al. Cerebral cavernous malformations. Incidence and familial occurrence. N. Engl. J. Med. 319, 343–347 (1988).
Dogruluk, T. et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 75, 5341–5354 (2015).
Gault, J., Shenkar, R., Recksiek, P. & Awad, I. A. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke 36, 872–874 (2005).
Akers, A. L., Johnson, E., Steinberg, G. K., Zabramski, J. M. & Marchuk, D. A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 18, 919–930 (2009).
McDonald, D. A. et al. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum. Mol. Genet. 23, 4357–4370 (2014).
Xu, L. et al. Clonal evolution and changes in two AML patients detected with a novel single-cell DNA sequencing platform. Sci. Rep. 9, 11119 (2019).
Lopez-Ramirez, M. A. et al. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. J. Exp. Med. 214, 3331–3346 (2017).
Marchi, S. et al. Defective autophagy is a key feature of cerebral cavernous malformations. EMBO Mol. Med. 7, 1403–1417 (2015).
Abdulrauf, S. I., Kaynar, M. Y. & Awad, I. A. A comparison of the clinical profile of cavernous malformations with and without associated venous malformations. Neurosurgery 44, 41–46, discussion 46–47 (1999).
Tan, W. H. et al. The spectrum of vascular anomalies in patients with PTEN mutations: implications for diagnosis and management. J. Med. Genet. 44, 594–602 (2007).
Adams, D. M. et al. Efficacy and safety of sirolimus in the treatment of complicated vascular anomalies. Pediatrics 137, e20153257 (2016).
Ozeki, M. et al. The impact of sirolimus therapy on lesion size, clinical symptoms, and quality of life of patients with lymphatic anomalies. Orphanet J. Rare Dis. 14, 141 (2019).
Triana, P. et al. Sirolimus in the treatment of vascular anomalies. Eur. J. Pediatr. Surg. 27, 86–90 (2017).
Venot, Q. et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 558, 540–546 (2018).
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483-486 (2010).
Ridder, D. A. et al. TAK1 in brain endothelial cells mediates fever and lethargy. J. Exp. Med. 208, 2615–2623 (2011).
Claxton, S. et al. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis 46, 74–80 (2008).
Chan, A. C. et al. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J. Clin. Invest. 121, 1871–1881 (2011).
Foster, K. W. et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24, 1491–1500 (2005).
Trotman, L. C. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003).
Adams, J. R. et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 71, 2706–2717 (2011).
Körbelin, J. et al. A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol. Med. 8, 609–625 (2016).
Hurst, C. D., Zuiverloon, T. C., Hafner, C., Zwarthoff, E. C. & Knowles, M. A. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res. Notes 2, 66 (2009).
Martelotto, L. G. ‘Frankenstein’ protocol for nuclei isolation from fresh and frozen tissue for snRNAseq. protocols.io https://doi.org/10.17504/protocols.io.3fkgjkw (2020).
Acknowledgements
We thank the members of the Kahn laboratory for thoughtful comments and advice; A. Alliance for enrolling patients; the University of Chicago PaleoCT core facility for its expertise in imaging and image quantification; the Penn CDB Microscopy Core for support with microscopy; Duke University School of Medicine for use of the Sequencing and Genomic Technologies Shared Resource for library preparation and sequencing; and K. Wood (Duke University) for providing the cell-line DNA used as a positive control in the ddPCR assay design. Flow cytometry was performed in the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility. These studies were supported by National Institute of Health grants R01HL094326 and R01NS100949 (to M.L.K.), P01NS092521 (M.L.K., D.A.M. and I.A.A.) the Leducq Foundation (M.L.K. and M.P.), the AHA-Allen foundation (M.L.K.), T32 HL007150 (A.A.R.), F31HL152738 (D.A.S.), F31NS115256 (C.C.H.) and F30NS100252 (A.T.T.); European Research Council (ERC) Synergy grant-2019-WATCH-810331 (to M.S.); and ERC Consolidator grant EMERGE-773047 (to M.P.).
Author information
Authors and Affiliations
Contributions
A.A.R. designed and performed most of the mouse and tissue culture experiments and wrote the manuscript. D.A.S. carried out the genetic studies of human CCM lesions and wrote the manuscript. Y.S.S. created and carried out the cranial window assays in adult mice. C.C.H., A.T.T., M.R.D. and H.M.-F. contributed to mouse genetic studies. M. Castro and M. Chen performed in vitro studies. N.H., R.G., S.R., R.L., T.M., R.S. and I.A.A. carried out microCT imaging and quantification of CCM lesions in a blinded manner. M. Chen, C.B. and P.M. assisted with mouse genetic studies. M.M.B. assisted with snDNA-seq studies. J.Y. and D.C.S. carried out histological studies. M.T.L. provided surgically excised human samples. M.R.D., M.S., J.M.R. and J.K. provided critical reagents. M.P., I.A.A., D.A.M. and M.LK. designed experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests. I.A.A. is Chairman of the Scientific Advisory Board for Angioma Alliance and provides expert opinions related to clinical care of cerebral cavernous malformations.
Additional information
Peer review information Nature thanks Marcus Goncalves, Mustafa Sahin, Salim Seyfried and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
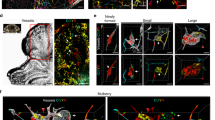
Extended Data Fig. 1 Loss of CCM gene function in endothelial cells in adult mice confers cavernous vascular malformations in the testis but not the brain.
a, Experimental design for investigating the effects of deleting CCM gene activity in neonatal endothelial cells. b, Cavernous malformations form by P10 in the hindbrain of Krit1iECKO animals with a susceptible gut microbiome. Shown are images of hindbrains from the indicated animals (top panels), and haematoxylin-and-eosin (H&E)-stained histological sections (lower four panels). The arrow indicates a CCM lesion in the venous vessel of the white matter. c, Schematic showing deletion of CCM genes in endothelial cells (ECs) of adult mice on a susceptible microbiome background. d, Cavernous malformations are not detected in the brains of six-month-old Krit1iECKO animals following administration of tamoxifen. Images of hindbrains from the indicated animals are shown above, and H&E-stained histological sections below. e, Cavernous malformations are detected in the testes of six-month-old Krit1iECKO animals. Images of testis from the indicated animals are shown above, and H&E-stained histological sections below. Asterisks indicate blood-filled testes. Arrows indicate cavernous blood-filled vessels around the seminiferous tubules. In b, d, e, visual images are representative of n = 4 animals per genotype; H&E histology is representative of six tissue sections from n = 4 animals. Scale bars for visual images, 1 mm; scale bars for histology, 0.1 mm. f, Immunostaining for KLF4 and the endothelial-cell marker PECAM1 in brain (top) and testis (bottom) from the experiment in c. Note that endothelial CCM LOF in adult mice results in upregulation of KLF4 without CCM formation in the brain. Arrows indicate KLF4-positive nuclei in PECAM1-positive ECs. Yellow arrowheads indicate KLF4-positive peritubular myoid cells. Scale bars, 50 μm. g, Quantification of KLF4-positive and KLF4-negative ECs identified using co-staining for KLF4 and PECAM1 in testis. Quantification from ten individual 800 μm × 800 μm high-power fields from three individual animals. h, Immunostaining for DPEAAE, a versican neo-epitope exposed by ADAMTS-mediated proteolysis, in Krit1iECKO testis. Arrows indicate peri-endothelial cell detection of DPEAAE around testicular cavernomas. Scale bars, 0.1 mm. The control animals in b, d–g had genotypes of either Cdh5–CreERT2;Krit1fl/+ or Krit1fl/fl. Immunofluorescence images in f, h are representative of six tissue sections from n = 4 individual animals per genotype.
Extended Data Fig. 2 Vascular lesions due to CCM LOF and/or PIK3CA GOF arise in veins of the white matter.
a, Neonatal endothelial induction of Krit1 deletion and/or PIK3CAH1047R expression. b, c, H&E staining of saggital hindbrain sections from P6 control, Pik3caiBECGOF, Krit1iBECKO;Pik3caiBECGOF (b and b′) and Krit1iBECKO (c and c′) animals with a resistant (Res) or susceptible (Susc) microbiome. Samples in b′, c′ were obtained from animals distinct from those in b, c. Note that lesions form in the white matter with both CCM LOF and PIK3CA GOF. Boxes in the upper images of each panel denote the area of the magnified image immediately below. Dotted lines outline the white matter of the cerebellum. Arrows indicate lesions in the veins and venules of the white matter. H&E images are representative of six tissue sections from n = 4 animals per genotype. wm, white matter. Scale bars, 0.1 mm.
Extended Data Fig. 3 Endothelial Pten LOF synergizes with Krit1 LOF in a dose-dependent manner.
a, Tamoxifen (TMX)-induced deletion of Krit1 and of either none or one allele of Pten in neonatal endothelial cells, using a Pdgfb–CreERT2 transgene. b, Representative visual (top) and paired microCT (bottom) images of Krit1iECKO and Krit1iECKO;Ptenfl/+ littermate mice on a susceptible microbiome background at P12. Scale bars, 1 mm. These mice were produced from a Ptenfl/+ by Ptenfl/+ cross; however, no Krit1iECKO;Ptenfl/fl littermates survived to P12. c, MicroCT-based quantification of lesion volumes at P12. Krit1iECKO, n = 6; Krit1iECKO;Ptenfl/+, n = 13. d, Induction of Krit1 deletion and deletion of either one or both alleles of Pten in neonatal endothelial cells using a Pdgfb–CreERT2 transgene with brains harvested at P7. These mice were produced from a Ptenfl/+ by Ptenfl/fl cross. e, Representative visual and paired microCT images in Krit1iECKO;Ptenfl/+ and Krit1iECKO;PteniECKO littermate mice at P7. Scale bars, 1 mm. f, MicroCT quantification of lesion volumes at P7. Krit1iECKO;Ptenfl/+, n = 13; Krit1iECKO;PteniECKO, n = 8. Data are means ± s.e.m. Unpaired, two-tailed Welch’s t-test.
Extended Data Fig. 4 Uninduced Slco1c1–CreERT2;Krit1fl/fl;iPik3caH1047R animals develop focal lesions owing to endothelial leakage of the Slco1c1–CreERT2 transgene.
a, Generation of a survival curve in the absence of tamoxifen administration. b, Postnatal survival curves in the absence of tamoxifen administration for the indicated genotypes. Slco1c1–CreERT2;Krit1fl/fl; n = 15, Slco1c1–CreERT2;Krit1fl/+;iPik3caH1047R, n = 10; Slco1c1–CreERT2;Krit1fl/fl;iPik3caH1047R, n = 39. Log-rank test. c, Representative visual and paired microCT images of brains harvested from untreated P28 littermates. Scale bars, 1 mm. d, MicroCT quantification of lesion volumes of untreated P28 animals. Slco1c1–CreERT2;Krit1fl/fl, n = 5; Slco1c1–CreERT2;Krit1fl/+;iPik3caH1047R, n = 9; Slco1c1–CreERT2;Krit1fl/fl;iPik3caH1047R n = 10. Data are shown as means ± s.e.m. Unpaired, two-tailed Welch’s t-test. e, Additional visual images of brains from a superior and inferior perspective from animals harvested at various time points (P19 to P36). Arrows point to focal vascular lesions. Scale bars, 1 mm. f, Leak assessed by immunostaining of brain sections with antibodies against GFP to identify Cre-expressing cells, and against cell-surface marker PECAM1 (top) and nuclear protein ERG (bottom) to identify endothelial cells. Scale bars, 50 μm. Immunofluorescence images are representative of ten tissue sections from n = 4 individual animals per genotype. g, Quantitation of GFP- and ERG-positive nuclei (n = 242) and GFP-positive, ERG-negative nuclei (n = 3) from 20 individual 800 μm × 800 μm high-power fields.
Extended Data Fig. 5 Exogenous delivery of Cre recombinase via AAV vector to drive combined loss of CCM function and gain of PIK3CA function results in formation of CCMs in the adult brain.
a, Experimental approach in which a cranial window is created and AAV–Cre virus injected into the brains of 2-month-old mice, with serial imaging at post-operative days 1, 7, 10, 14, 18 and 21. b, Representative visual images of brains harvested 21 days after injection of AAV–Cre into adult animals. Dotted circles indicate the site of the cranial window and AAV–Cre injection. This panel includes the visual images displayed in Fig. 1. Scale bars, 1 mm. c, Serial images obtained through the cranial window of mice of the indicated genotypes following injection of AAV–Cre. The iPik3caH1047R designation includes iPik3caH1047R and/or Krit1fl/+;iPik3caH1047R genotypes. White arrows indicate cavernous malformations in Krit1fl/fl;iPik3caH1047R mice. Black arrows indicate hypervascularity in iPik3caH1047R mice. d, Staining for perilesional iron deposition in brains, indicative of chronic bleeding, from four independent Krit1fl/fl;iPik3caH1047R mice at post-operative day 21. Scale bars, 200 μm.
Extended Data Fig. 6 Lineage tracing of AAV–Cre activity after direct injection into the mouse brain.
AAV–Cre was injected into the brain of Ai14 Cre reporter animals, and Cre activity was assessed by detection of the tdTomato (red fluorescent protein, RFP) reporter 14 days after injection. a–c, Confocal microscopic overviews of: a, the injection site; b, the border region of viral spread; and c, the contralateral cortex of AAV–Cre-injected Ai14 mice, two weeks after stereotactic injection. AAV–Cre-transduced cells expressed RFP (shown in white). RFP-positive vessels were identified by colocalization with PECAM1 (red). White arrows point to representative RFP-positive vessels. Yellow arrowheads point to RFP-expressing neuronal cells. Scale bars, 100 μm. The numbered boxed regions are shown at higher magnification on the right: 1 and 2 show anti-PECAM1 staining for endothelial cells, overlaid with RFP signal; 1′ and 2′ show PECAM staining alone; 1′′ and 2′′ shown RFP staining alone. These data are representative of 12 separate images from 8 tissue sections from n = 2 individual animals. Scale bar, 20 μm.
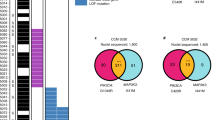
Extended Data Fig. 7 Characterization of snDNA-seq of human CCM samples.
a, Relationship between somatic PIK3CA and CCM mutations detected in bulk sequencing. Points indicate individual mutations in either a CCM gene or PIK3CA. Lines connect the CCM gene and PIK3CA mutations present in a single sample. Box plots show the aggregate frequencies of PIK3CA and CCM mutations. Centre lines show medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th to 75th percentiles; outliers are represented by dots. n = 21 sample points for both plots. b, Representative FANS plots of unstained (top row) and DAPI-stained (bottom row) CCM homogenates. Doublets are discriminated by forward scatter profiles for DAPI-stained samples. FSC, forward scatter; SSC, side scatter. Plots consist of 100,000 events. The unstained sample contains one event (0%) in the DAPI-positive singlet gate. The DAPI-stained samples contain 2,414 events (2.4%) in the DAPI-positive singlet gate. c, Total reads and average coverage per nucleus from snDNA-seq for each mutation detected by bulk sequencing. The dotted line shows 20× coverage, the minimum cutoff used for establishing genotype. d, Pseudobulk allele frequency from snDNA-seq for each mutation detected by bulk sequencing. The dotted line shows an allele frequency of 1%. VAF, variant allele frequency. Note that the data point with the arrow in c, d shows a mutation in sample 5079 detected in bulk sequencing, which, owing to poor amplification during snDNA-seq, received insufficient coverage per nucleus (4.5×) to establish nuclear genotypes, but is clearly present in pseudobulk reads (1,849 of 9,814). e, Comparison of mutation allele frequency as detected by bulk sequencing and snDNA-seq. As nuclei are diploid for the relevant autosomes, the x-axis is equal to the fraction of mutant nuclei divided by two. The dotted line shows perfect correlation at x = y. R and P were calculated by Pearson’s correlation coefficient. f, A summary of snDNA-seq results for three sporadic and two familial CCMs. The number of nuclei with each possible genotype are listed. Plus symbols indicate wild-type alleles; asterisks indicate mutant alleles. Note that only one somatic CCM mutation was identified in samples 5038 and 5079. P values were determined by two-tailed chi-squared tests between the observed and expected triple-mutant nuclei (or double-mutant for lesions 5038 and 5079) determined by Poisson distribution (see Methods).
Extended Data Fig. 8 PI3K signalling does not augment MEKK3–KLF2/4 signalling.
a, How gain of PI3K signalling might augment CCM formation by acting upstream of MEKK3–KLF2/4 signalling in endothelial cells. b, Immunostaining for KLF4 and PECAM1 in hindbrain sections from P6 control, Krit1BECKO, Pik3caiBECGOF and Krit1iBECKO;Pik3caiBECGOF neonates with either a susceptible (Susc) or resistant (Res) gut microbiome. White arrows indicate endothelial-cell nuclear KLF4 staining. Immunofluorescence images are representative of ten tissue sections from n = 4 individual animals per genotype. Control animals are either Slco1c1–Cre;Krit1fl/+ or Krit1fl/fl. Scale bars, 50 μm. c, Measurement of Klf2 and Klf4 mRNA in endothelial cells isolated from the hindbrains of P6 control, Krit1BECKO and Pik3caiBECGOF neonates. Control, n = 8; Krit1BECKO, n = 6; Pik3caiBECGOF, n = 8. d, Measurement of KLF2 and KLF4 mRNA in HUVECs treated with the indicated siRNAs or lentiviral vectors. n = 6 individual wells per group over two independent experiments. Data are shown as means ± s.e.m. Unpaired, two-tailed Welch’s t-test.
Extended Data Fig. 9 The CCM effector KLF4 augments endothelial cell PI3K-mTORC1 signalling.
a, Neonatal endothelial induction of KLF4 expression in KLF4iBECGOF animals. b, Immunostaining for KLF4 and PECAM1 in hindbrain sections from P6 control and KLF4iBECGOF animals. Boxes in the upper images denote the area of the magnified images immediately below. Immunofluorescence images are representative of six tissue sections from n = 4 individual animals per genotype. Scale bars, 50 μm. hKLF4, hindbrain KLF4. c, H&E-stained sections of hindbrain from control and KLF4iBECGOF littermates. Boxes in the upper images denote the area of the magnified images immediately below. Black arrows indicate lesions. Dotted lines outline the white matter (wm) of the cerebellum. Note that the dilated white-matter venules are similar to those observed with CCM LOF and PIK3CA GOF in Extended Data Fig. 2. The H&E histology is representative of six tissue sections from n = 4 animals per genotype. Scale bars, 0.1 mm. d, Immunostaining for phosphorylated S6 ribosomal protein (p-S6) and PECAM1 in hindbrain sections from P6 control and KLF4iBECGOF animals. White and yellow arrows indicate p-S6-positive endothelial and nonendothelial cells, respectively. Immunofluorescence images are representative of six tissue sections from n = 4 individual animals per genotype. Scale bars, 50 μm. e, Immunoblot detection of KLF4, KLF4–GFP and eNOS (whose gene is a KLF4 target) in HUVECs without and with inducible lentiviral expression of KLF4–GFP (iKLF4 cells) or control lentivirus. Tubulin is shown as a loading control.
Extended Data Fig. 10 Rapamycin-mediated rescue of CCM formation is independent of KLF4.
a, Experimental approach in which a cranial window is created and AAV–Cre is injected into the brains of two-month-old mice, followed by daily injection of vehicle, 100 μg of rapamycin (low) or 400 μg of rapamycin (high) from post-operative day 7 to day 21, with serial imaging at post-operative days 1, 7, 10, 14, 18 and 21. b, Representative visual images of brains harvested 21 days after injection of AAV–Cre and 2 weeks of daily vehicle or rapamycin treatment in Krit1fl/fl;iPik3caH1047R mice. Dotted circles indicate the site of cranial window and AAV–Cre injection. Arrows indicate detached lesions. Scale bars, 1 mm. c, Serial images were obtained through the cranial window of Krit1fl/fl;iPik3caH1047R mice following injection of AAV–Cre and subsequent treatment. Arrows indicate the formation and growth of individual cavernous malformations. d, MicroCT quantification of lesion volumes 21 days after creation of the cranial window and injection of AAV–Cre. The values shown include those from Fig. 4 (vehicle and low-dose treatments). Vehicle, n = 6; rapamycin low, n = 7; rapamycin high, n = 7. Data are shown as means ± s.e.m. Unpaired, two-tailed Welch’s t-test. e, Neonatal endothelial induction of Krit1 deletion and treatment with rapamyin or vehicle control at P2. f, Immunostaining for PECAM1 and p-S6 of hindbrain sections from P6 Krit1iBECKO animals treated with vehicle or rapamycin. White arrows indicate p-S6-positive endothelial cells, seen in the control but not rapamycin-treated animals. Yellow arrowheads indicate p-S6-positive neuronal cells. g, Immunostaining ofor PECAM1 and KLF4 of hindbrain sections from P6 Krit1iBECKO animals treated with vehicle or rapamycin. White arrows indicate KLF4-positive endothelial cells, detected in control and rapamycin-treated animals. Scale bars, 50 μm.
Supplementary information
Supplementary Figure 1
This file contains the uncropped gels.
Supplementary Table 1
Human CCM bulk tissue mutation data. Mutation data for each human CCM analyzed in this study using targeted sequencing, ddPCR, or SNaPshot. Each sample is notated as either familial (F), sporadic (S), or unknown (blank) as well as the affected gene for familial samples (CCM1/2/3). ddPCR and SNaPshot assay results for PIK3CA E542K, E545K, and H1047R for each sample are listed including the detected allele frequency as well as final mutation call. SNaPshot calls are color coded by variant and ddPCR results are colored according to the detected allele frequency for each variant. Sequencing data for PIK3CA are reported as the number of alt and ref reads supporting the listed variant, the allele frequency, and the final mutation call. PIK3CA variants shown in red text denote samples with insufficient coverage to make a definitive variant call per the thresholds detailed in the methods section; variant calls for these samples rely on a positive result by ddPCR and confirmation by a tertiary assay (SNaPshot). Germline and somatic mutations in the CCM genes are reported in respective columns listing the chromosomal coordinates in hg19, the predicted functional consequence of the variant, the number of reads supporting the alt and ref alleles, and the resulting allele frequency. The functional consequence of missense and in-frame indels are predicted by in silico tools SIFT (scale: 0-1, higher is more damaging) and PROVEAN (scale: continuous, <-2.5 predicted damaging) where available.
Supplementary Table 2
Human CCM snDNA-seq read counts. Number of total, mutant, and wild type supporting reads present in each nucleus for PIK3CA and CCM mutations. Each nucleus is denoted by an 18bp oligo barcode which is incorporated into each read to define the nucleus of origin. These data were processed to generate Fig 2e. and Extended Data Figure 7c-f.
Rights and permissions
About this article
Cite this article
Ren, A.A., Snellings, D.A., Su, Y.S. et al. PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature 594, 271–276 (2021). https://doi.org/10.1038/s41586-021-03562-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03562-8
This article is cited by
-
mTOR inhibitor reduces nontumour-related death in liver transplantation for hepatocellular carcinoma
Molecular Biomedicine (2024)
-
Discovery and Characterization of Ephrin B2 and EphB4 Dysregulation and Novel Mutations in Cerebral Cavernous Malformations: In Vitro and Patient-Derived Evidence of Ephrin-Mediated Endothelial Cell Pathophysiology
Cellular and Molecular Neurobiology (2024)
-
Transcriptomic signatures of individual cell types in cerebral cavernous malformation
Cell Communication and Signaling (2024)
-
Ageing as a software design flaw
Genome Biology (2023)
-
Effective long-term sirolimus treatment in hypoxemia mainly due to intrapulmonary right-to-left shunt in a patient with multiple vascular anomalies
Orphanet Journal of Rare Diseases (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.