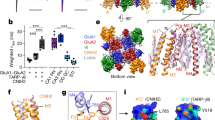
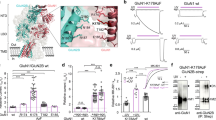
Abstract
AMPA-selective glutamate receptors mediate the transduction of signals between the neuronal circuits of the hippocampus1. The trafficking, localization, kinetics and pharmacology of AMPA receptors are tuned by an ensemble of auxiliary protein subunits, which are integral membrane proteins that associate with the receptor to yield bona fide receptor signalling complexes2. Thus far, extensive studies of recombinant AMPA receptor–auxiliary subunit complexes using engineered protein constructs have not been able to faithfully elucidate the molecular architecture of hippocampal AMPA receptor complexes. Here we obtain mouse hippocampal, calcium-impermeable AMPA receptor complexes using immunoaffinity purification and use single-molecule fluorescence and cryo-electron microscopy experiments to elucidate three major AMPA receptor–auxiliary subunit complexes. The GluA1–GluA2, GluA1–GluA2–GluA3 and GluA2–GluA3 receptors are the predominant assemblies, with the auxiliary subunits TARP-γ8 and CNIH2–SynDIG4 non-stochastically positioned at the B′/D′ and A′/C′ positions, respectively. We further demonstrate how the receptor–TARP-γ8 stoichiometry explains the mechanism of and submaximal inhibition by a clinically relevant, brain-region-specific allosteric inhibitor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM maps and coordinates for overall, the ATD layer and the LBD–TMD layer of the A1A2A1A2 symmetric (S) and A1A2A1A2 asymmetric (AS) complexes have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-23283 and EMD-23284 and in the Protein Data Bank (PDB) under accession codes 7LDD and 7LDE, respectively. The cryo-EM maps for overall, the ATD layer and the LBD–TMD layer of the A1A2A3A2 (AS1), A1A2A3A2 (AS2), A3A2A3A2 (S) and A3A2A3A2 (AS) complexes have been deposited in the EMDB under accession numbers EMD-23285, EMD-23286, EMD-23287 and EMD-23288, respectively. The cryo-EM maps of A1A2A3A2 (AS3) and A1A2AXA2 have been deposited in the EMDB under accession numbers EMD-23289 and EMD-23290, respectively. The cryo-EM map and coordinates for the LBD-TMDmix complex have been deposited in the EMDB and PDB under accession codes EMD-23292 and 7LEP, respectively.
References
Diering, G. H. & Huganir, R. L. The AMPA receptor code of synaptic plasticity. Neuron 100, 314–329 (2018).
Jackson, A. C. & Nicoll, R. A. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 (2011).
Collingridge, G. L., Kehl, S. J. & McLennan, H. Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. J. Physiol. 334, 33–46 (1983).
Bliss, T. V. & Lomo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 331–356 (1973).
Rosenmund, C., Stern-Bach, Y. & Stevens, C. F. The tetrameric structure of a glutamate receptor channel. Science 280, 1596–1599 (1998).
Sobolevsky, A. I., Rosconi, M. P. & Gouaux, E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009).
Traynelis, S. F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010).
Keinanen, K. et al. A family of AMPA-selective glutamate receptors. Science 249, 556–560 (1990).
Schwenk, J. et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84, 41–54 (2014).
Wenthold, R. J., Petralia, R. S., Blahos J, I. I. & Niedzielski, A. S. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J. Neurosci. 16, 1982–1989 (1996).
Kamalova, A. & Nakagawa, T. AMPA receptor structure and auxiliary subunits. J. Physiol. 599, 453–469 (2021).
Tomita, S. et al. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J. Cell Biol. 161, 805–816 (2003).
Schwenk, J. et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319 (2009).
Chen, L. et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943 (2000).
Herguedas, B. et al. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Science 364, eaav9011 (2019).
Nakagawa, T. Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366, 1259–1263 (2019).
Jain, A. et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011).
Zhao, Y., Chen, S., Swensen, A. C., Qian, W. J. & Gouaux, E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362 (2019).
Maher, M. P. et al. Discovery and characterization of AMPA receptor modulators selective for TARP-γ8. J. Pharmacol. Exp. Ther. 357, 394–414 (2016).
Lu, W. et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62, 254–268 (2009).
Jacobi, E. & von Engelhardt, J. Diversity in AMPA receptor complexes in the brain. Curr. Opin. Neurobiol. 45, 32–38 (2017).
Morise, J. et al. Distinct cell surface expression patterns of N-glycosylation site mutants of AMPA-type glutamate receptor under the homo-oligomeric expression conditions. Int. J. Mol. Sci. 21, 5101 (2020).
Fukaya, M. et al. Abundant distribution of TARP γ-8 in synaptic and extrasynaptic surface of hippocampal neurons and its major role in AMPA receptor expression on spines and dendrites. Eur. J. Neurosci. 24, 2177–2190 (2006).
Rouach, N. et al. TARP γ-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat. Neurosci. 8, 1525–1533 (2005).
Kato, A. S. et al. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096 (2010).
Carrillo, E. et al. Mechanism of modulation of AMPA receptors by TARP-γ8. J. Gen. Physiol. 152, jgp.201912451 (2020).
Plested, A. J. & Mayer, M. L. AMPA receptor ligand binding domain mobility revealed by functional cross linking. J. Neurosci. 29, 11912–11923 (2009).
Baranovic, J. et al. Dynamics of the ligand binding domain layer during AMPA receptor activation. Biophys. J. 110, 896–911 (2016).
Harmel, N. et al. AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS ONE 7, e30681 (2012).
Boudkkazi, S., Brechet, A., Schwenk, J. & Fakler, B. Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron 82, 848–858 (2014).
Gill, M. B., Kato, A. S., Wang, H. & Bredt, D. S. AMPA receptor modulation by cornichon-2 dictated by transmembrane AMPA receptor regulatory protein isoform. Eur. J. Neurosci. 35, 182–194 (2012).
Herring, B. E. et al. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77, 1083–1096 (2013).
Sommer, B., Köhler, M., Sprengel, R. & Seeburg, P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 (1991).
Burnashev, N., Villarroel, A. & Sakmann, B. Dimensions and ion selectivity of recombinant AMPA and kainate receptor channels and their dependence on Q/R site residues. J. Physiol. 496, 165–173 (1996).
Swanson, G. T., Kamboj, S. K. & Cull-Candy, S. G. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J. Neurosci. 17, 58–69 (1997).
Bowie, D. & Mayer, M. L. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron 15, 453–462 (1995).
Lomeli, H. et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266, 1709–1713 (1994).
Kirk, L. M. et al. Distribution of the SynDIG4/proline-rich transmembrane protein 1 in rat brain. J. Comp. Neurol. 524, 2266–2280 (2016).
Matt, L. et al. SynDIG4/Prrt1 is required for excitatory synapse development and plasticity underlying cognitive function. Cell Rep. 22, 2246–2253 (2018).
Troyano-Rodriguez, E., Mann, S., Ullah, R. & Ahmad, M. PRRT1 regulates basal and plasticity-induced AMPA receptor trafficking. Mol. Cell. Neurosci. 98, 155–163 (2019).
Penn, A. C. et al. Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature 549, 384–388 (2017).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014).
Coleman, J. A., Green, E. M. & Gouaux, E. X-ray structures and mechanism of the human serotonin transporter. Nature 532, 334–339 (2016).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Sultan, F. A. Dissection of different areas from mouse hippocampus. Bio-Protocol 3, e955 (2013).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
The PyMOL Molecular Graphics System v.2.1 (Schrödinger, 2020).
Biedermann, J., Braunbeck, S., Plested, A. J. & Sun, H. Non-selective cation permeation in an AMPA-type glutamate receptor. Proc. Natl Acad. Sci. USA 118, e2012843118 (2021).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Jain, A., Liu, R., Xiang, Y. K. & Ha, T. Single-molecule pull-down for studying protein interactions. Nat. Protocols 7, 445–452 (2012).
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002).
Acknowledgements
We thank the Pacific Northwest Cryo-EM Center (PNCC) and OHSU MMC for microscope use, D. Cawley, P. Streeter, Y. Zhong and N. Sheldon for generating antibodies, A. Goehring for mouse dissections, M. Mayer for his guidance and advice on electrophysiology experiments, J. Elferich for help writing SiMPull processing scripts, S. Chen for help initiating the project, L. Vaskalis for assistance with figures, F. Jalali-Yazdi and H. Owen for help with manuscript preparation and R. Nicoll and members of the Gouaux laboratory for discussions. PNCC is supported by NIH grant U24GM129547 and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. T.H. and J.M. were supported by the NIGMS grant R35GM122569. This work was supported by the NINDS grant R01NS038631 to E.G. T.H. and E.G. are investigators of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institutes of Health.
Author information
Authors and Affiliations
Contributions
J.Y., P.R. and E.G. designed the project. J.Y. and P.R. performed the sample preparation for cryo-EM and biochemistry studies. J.Y. and P.R. performed the cryo-EM data collection, data analysis and model building. J.Y. performed the patch-clamp recording experiments. S.C. performed all of the SiMPull experiments with T.H and J.M. providing training and comments. E.G., J.Y., P.R. and S.C. wrote the manuscript with input from T.H. and J.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Sudha Chakrapani, Vasanthi Jayaraman and Andrew Plested for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Biochemical characterization and cryo-EM analysis of hpAMPAR complexes.
a, Representative SEC profile of hippocampal AMPAR complexes. Inset shows an SDS–PAGE gel of AMPAR complexes and antibody fragments used for cryo-EM grid preparation, visualized by silver staining. The gel was repeated three times from different batches of purification with similar results. b, Western blot analysis of isolated AMPAR complexes using antibodies against GluA1, GluA2, GluA3, GluA4, PSD95, TARP-γ8 and CNIH2. The uncropped blot can be found in Supplementary Fig. 1 and blotting was repeated three times with similar results. c, A representative cryo-EM micrograph of hpAMPAR complexes. The experiments were repeated four times with similar results. d, Selected two-dimensional class averages. Protrusions extending out of the detergent micelle are indicated by arrows, corresponding to the extracellular domain of TARP-γ8. Similar results were obtained from experiments repeated four times.
Extended Data Fig. 2 Characterization of monoclonal antibodies 13A8 and E3.
a, Octet measurements of the 13A8 monoclonal antibody binding to TARP-γ8. Concentrations of the 13A8 monoclonal antibody ranging from 25 nM to 200 nM were applied. b–f, FSEC profiles of recombinant GFP-tagged TARP-γ8 (b), TARP-γ2 (c), TARP-γ3 (d), TARP-γ4 (e) and TARP-γ7 (f) with 13A8 monoclonal antibody (green traces) and without 13A8 monoclonal antibody (black traces), detecting GFP fluorescence. Only the TARP-γ8 trace is shifted by the 13A8 monoclonal antibody. g, Octet measurements of the E3 monoclonal antibody binding to GluA4. h–k, FSEC profiles of recombinant mKalama-tagged GluA1 (h), GFP-tagged GluA2 (i), GFP-tagged GluA3 (j) and GFP-tagged GluA4 (k) with E3 monoclonal antibody (green traces) and without E3 monoclonal antibody (black traces), detecting mKalama or GFP fluorescence. Only GluA4 receptors are shifted by the E3 monoclonal antibody.
Extended Data Fig. 3 A representative flow chart of data processing focused on the whole receptor and ATD layer using data-processing strategy 1.
A total of 2,893,667 particles was picked from 46,927 motion-corrected micrographs in cryoSPARC v.2.14. Classes showing clear receptor features were kept after several rounds of two-dimensional classification, resulting in the retention of 2,893,667 particles. Next, three-dimensional classification with a large sampling degree was performed to further remove junk classes in RELION 3.0. Another round of three-dimensional classification was carried out to sort receptors with the same Fab and scFv combination. Classes with the same ATD labelling and orientation were combined and subjected to ATD-focused classification without alignment. For the A1A2A1A2 symmetric subtype, the ATD layer was classified into eight classes, of which one class, which occupied the largest population (55%), had the least well-resolved secondary structure features. Another round of ATD-focused classification was performed on this class, producing a subtype with one unlabelled subunit, denoted as A1A2AXA2. The three remaining classes showing the most well-defined secondary structure features were selected for final refinement with C2 symmetry, producing a map at a resolution of 4 Å. ATD-focused refinement with C2 symmetry was carried out to improve map density in the ATD, yielding an ATD–A1A2A1A2 symmetric map at a resolution of 3.4 Å.
Extended Data Fig. 4 Data-processing workflow to determine AMPAR subtypes using data-processing strategy 2.
Motion-corrected micrographs were first curated on the basis of ice thickness, motion correction, CTF fit and astigmatism. Template-based picking was used to autopick 6,002,517 particles in cryoSPARC v.2.14. To remove junk particles and false positives, multiple rounds of two-dimensional and three-dimensional classification were performed, selecting only classes that showed discernible receptor features, resulting in a particle stack of 1,844,956 particles. To sort receptors based on subtype (AMPAR subunit stoichiometry and tilting), multiple rounds of three-dimensional classification were performed without symmetry imposed or masking. Particles from classes showing clear labelling with antibodies were grouped into distinct subtypes. Each of the AMPAR subtypes were refined separately. This strategy elucidated three different heteromeric AMPAR subtypes comprising both symmetric and asymmetric conformations.
Extended Data Fig. 5 Three-dimensional reconstructions of dimeric GluA1–GluA2 and dimeric GluA2–GluA3 complexes.
a, c, e, g, i, Local resolution estimates of the entire GluA1–GluA2 symmetric map (a), ATD layer of the GluA1–GluA2 symmetric map (c), entire GluA1–GluA2 asymmetric map (e), the ATD layer of GluA1–GluA2 asymmetric map (g) and the LBD–TMD layers of the GluA1–GluA2 map (i). d, h, k, FSC curves before and after masking and between the model and the final maps of the ATD layer of the GluA1–GluA2 symmetric map (d), the ATD layer of the GluA1–GluA2 asymmetric map (h) and the LBD–TMD layers of the GluA1–GluA2 map (k). j, Angular distribution of the LBD–TMD layers of the GluA1–GluA2 map. l, n, p, r, t, Local resolution estimates of the entire GluA2–GluA3 symmetric map (l), the ATD layer of the GluA2–GluA3 symmetric map (n), the entire GluA2–GluA3 asymmetric map (p), the ATD layers of the GluA2–GluA3 asymmetric map (r) and LBD–TMD layers of the GluA2–GluA3 map (t). b, f, m, o, q, s, v, FSC curves before and after masking of the whole GluA1–GluA2 symmetric map (b), the entire GluA1–GluA2 asymmetric map (f), the entire GluA2–GluA3 symmetric map (m), the ATD layer of GluA2–GluA3 symmetric map (o), the entire GluA2–GluA3 asymmetric map (q), the ATD layer of the GluA2–GluA3 asymmetric map (s) and the LBD–TMD layers of the GluA2–GluA3 map (v). u, Angular distribution of the LBD–TMD layers of the GluA2–GluA3 map.
Extended Data Fig. 6 Three-dimensional reconstructions of trimeric GluA1–GluA2–GluA3 complexes and the LBD–TMDmix map.
a, c, e, g, i, l, o, Local resolution estimates of the entire GluA1–GluA2–GluA3 asymmetric 1 map (a), the ATD layer of the GluA1–GluA2–GluA3 asymmetric 1 map (c), the entire GluA1–GluA2–GluA3 asymmetric 2 map (e), the ATD layer of the GluA1–GluA2–GluA3 asymmetric 2 map (g), the LBD–TMD layers of GluA1–GluA2–GluA3 without symmetry (i), the LBD–TMD layers of GluA1–GluA2–GluA3 with C2 symmetry imposed (l), and the LBD–TMDmix map (o). b, d, f, h, k, n, FSC curves before and after masking the entire GluA1–GluA2–GluA3 asymmetric 1 receptor map (b), the ATD layer of the GluA1–GluA2–GluA3 asymmetric 1 map (d), the entire GluA1–GluA2–GluA3 asymmetric 1 map (f), the ATD layer of the GluA1–GluA2–GluA3 asymmetric 2 map (h), the LBD–TMD layers of GluA1–GluA2–GluA3 map without symmetry (k) and with C2 symmetry (n). j, m, p, Angular distribution of the LBD–TMD layers of the GluA1–GluA2–GluA3 maps with C1 symmetry (j) or C2 symmetry (m), and the LBD–TMDmix map (p). q, FSC curves before and after masking and between the model and the final maps of the LBD-TMDmix map.
Extended Data Fig. 7 Representative TIRF images for native AMPAR complexes captured with the 15F1 monoclonal antibody.
a–e, Fluorescence detection with the anti-GluA1-Alexa488 monoclonal antibody (αGluA1) and anti-GluA3-Alexa594 monoclonal antibody (αGluA3) (a), the anti-GluA1-Alexa488 monoclonal antibody, anti-GluA3-Alexa594 monoclonal antibody and anti-GluA4-Alexa594 monoclonal antibody (αGluA4) (b), the anti-GluA1-Alexa488 monoclonal antibody, anti-GluA3-Alexa594 monoclonal antibody and anti-TARP-γ8 monoclonal antibody (αTARP-γ8) (for each colocalization experiment) (c), anti-TARP-γ8 (αTARP-γ8) Fab–GFP (d) and the anti-SynDIG4–Alexa594 monoclonal antibody (αSynDIG4) and anti-TARP-γ8–Alexa647 monoclonal antibody (αTARP-γ8) (e). Scale bars, 5 μm. For each SiMPull experiment, images were acquired from two independent samples on different days.
Extended Data Fig. 8 Structures of the dimeric GluA1–GluA2 receptor, trimeric GluA1–GluA2–GluA3 receptor and dimeric GluA2–GluA3 receptor complexes in symmetric and asymmetric conformations.
a, c, Cryo-EM structures of the GluA1–GluA2 subtype in symmetric (a) and asymmetric (c) conformations viewed parallel to the membrane. GluA1, GluA2, TARP-γ8 and CNIH2 are shown in grey, red, green and blue, respectively. Antibody fragments 11B8 scFv and 15F1 Fab are shown in pink and cyan, respectively. b, ATD layer analysis of symmetric and asymmetric conformations. Top, the ATD model of the symmetric state, in which the centre of mass (COM) of each subunit is indicated by a black circle. Bottom, the distances (in Ångstrom) and angles determined by the COMs of the symmetric (left) and asymmetric (right) conformations. d, e, Close contacts between the ATD layer and LBD layer in the asymmetric conformations. Magnified views of the ‘left’ side (d) and ‘right’ side (e) of the ATD–LBD interfaces as indicated in the black and cyan rectangles. f–h, Cryo-EM structures of the trimeric GluA1–GluA2–GluA3 subtype in asymmetric conformations with different tilted angles and orientations viewed parallel to the membrane. GluA1, GluA2, GluA3, TARP-γ8 and CNIH2 are coloured in grey, red, orange, green and blue, respectively. Antibody fragments 11B8 scFv, 15F1 Fab, 5B2 Fab are shown in pink, cyan and light yellow colours, respectively. i, j, Magnified views of ATD–LBD interfaces in the asymmetric states (f, h) as indicated in the black and red rectangles. The distances are defined by the Cα atoms of the indicated residues. k, l, Cryo-EM structures of the dimeric GluA2–GluA3 subtype in symmetric (k) and asymmetric (l) conformations viewed parallel to the membrane. m, The ATD layer analysis of the symmetric and asymmetric conformations. Top, an ATD model of the symmetric state. The COM of each subunit is shown as a black circle. Bottom, the distances (in Ångstrom) and angles determined by the COMs of the symmetric (left) and asymmetric (right) conformations. n, o, Close contacts between the ATD layer and LBD layer in the asymmetric conformations. Magnified views of the left side (n) and right side (o) of the ATD–LBD interfaces as indicated in the green and cyan rectangles.
Extended Data Fig. 9 Flow chart of data processing for hpAMPAR complexes focused on the LBD–TMD layers.
Particles corresponding to both the symmetric and the asymmetric GluA1–GluA2 subtypes were combined and subjected to LBD–TMD focused three-dimensional classification with alignment in RELION 3.0, resulting in three good classes with continuous transmembrane helical densities. Another round of classification without alignment was carried out for classes 1 and 8. Classes displaying strong density for TMD and auxiliary proteins were combined for refinement in cryoSPARC v.2.14, yielding the LBD–TMDA1/A2 map at a resolution of 3.63 Å.
Extended Data Fig. 10 Conformational differences in the LBD and TMD layers between native and recombinant AMPAR–auxiliary protein complexes.
a, Reference model and orientation of the hippocampal LBD–TMDA1/A2 complex. GluA1, GluA2, TARP-γ8 and CNIH2 are shown in grey, red, green and blue, respectively. b–e, Superposition of hippocampal LBD–TMDA1/A2 with recombinant GluA1–GluA2–TARP-γ8 complexes (PDB code: 6QKC) to show the differences in the LBD (b, d) and TMD (c, e) layers. Recombinant GluA1–GluA2–TARP-γ8 is shown in blue. COMs of LBD and TMD layers of each subunit are shown in coloured circles. The schematic diagrams illustrate the subunit arrangement differences in the distance (Ångstrom) of the LBD (d) and TMD (e) layers of these two complexes. f–i, Superposition of the hippocampal LBD–TMDA1/A2 structure with the recombinant GluA2–CNIH3 complex (PDB code: 6PEQ) to show the differences in the LBD (f, h) and TMD (g, i) layers. Recombinant GluA2–CNIH3 is shown in yellow. COMs of the LBD and TMD layers of each subunit are shown in coloured circles. The schematic diagrams illustrate the subunit arrangement differences in the distance (Ångstrom) of the LBD (h) and TMD (i) layers of these two complexes. j, The B/C LBD dimers from the hippocampal LBD–TMDA1/A2 structure and the GluA2–CNIH3 complex (PDB code: 6PEQ) were superimposed, exhibiting a 3.2 Å shift in the COM (black circles) between the opposing A/D LBD dimers. k, Superposition of the M1, M3 and M4 helices of the hippocampal LBD–TMDA1/A2 structure with the recombinant GluA2–CNIH3 complex (PDB code: 6PEQ), highlighting the rotation and compression of the GluA2–CNIH3 TMD layer. Equivalent positions of the Cα atoms from the M1 (Val538), M3 (Ile600) and M4 (Leu805) helices of the GluA2–CNIH3 structure are shifted by 4.5 Å, 5.7 Å and 4.7 Å, respectively.
Extended Data Fig. 11 Representative densities of the maps of the LBD–TMDA1/A2 or LBD–TMDmix complexes.
a, The S1–M1, M2–pore loop, R/G site and MPQX from GluA1 are isolated from LBD–TMDA1/A2, contoured at 0.085σ. b, S1–M1, M2–pore loop and R/G site from GluA2 are isolated from LBD–TMDA1/A2, contoured at 0.085σ. c, Comparison of the differences by fitting Arg and Gln into the GluA2 Q/R site density. d, Four transmembrane helices (TM1–TM4) in TARP-γ8 are isolated from LBD–TMDmix, contoured at 0.15σ. e, Four transmembrane helices (TM1–TM4) in CNIH2 are isolated from LBD-TMDmix, contoured at 0.13σ.
Extended Data Fig. 12 Electrophysiological recordings of GluA1–TARP-γ8 mutant proteins.
a, Current responses of wild-type GluA1–TARP-γ8 complexes evoked by repeated application of 10 mM glutamate with 10 pulses, each for a duration time of 1 s to reach a plateau of the steady-state current. To measure the inhibition of glutamate-induced currents, 10 μM JNJ-555511118 was applied before and during glutamate application for 1 s. Bottom insets illustrate the inhibition effect of JNJ-55511118 on the steady-state current by overlaying the currents without (the last application) and with JNJ-55511118 at timescales of 500 ms (left) and 20 ms (right). b–h, Representative recordings for the indicated GluA1 (b–e) and TARP-γ8 (f–h) mutant proteins with the same recording conditions as for the wild-type proteins.
Extended Data Fig. 13 LBD–TMDmix data-processing strategy 2.
Particles after two-dimensional and three-dimensional classification clean-up using data-processing strategy 2 were combined into a single stack and refined, and unless otherwise specified, all subsequent processing was performed in cryoSPARC v.2.14. Signal subtraction was implemented using the consensus refinement and a soft mask created around the ATD layer and all possible binding sites of the antibodies. Several rounds of two-dimensional classification were used to remove false positives and particles that still contained the ATD layer. This cleaned stack of particles underwent three-dimensional classification (C1 symmetry), which resulted in a single class displaying continuous transmembrane density features. Particles from this class were subject to two-dimensional classification to remove a small subset of junk particles. An iterative, sequential, refinement procedure consisting of (1) homogenous refinement, (2) non-uniform refinement, (3) local CTF refinement and (4) non-uniform refinement, was used to improve the resolution of the stack of 151,141 particles. This procedure was iterated twice until no resolution improvement was obtained, resulting in a 3.45 Å map. Particles from this map were then subjected to ab initio classification permitting the removal of junk particles. A new stack of 132,427 particles was then subjected to the previously described four-step refinement procedure for one iteration, before three-dimensional classification was performed in RELION 3.0 to remove junk particles. This final particle stack was subjected to non-uniform refinement in cryoSPARC to obtain the LBD–TMDmix map at 3.25 Å.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Tables 1-2.
Rights and permissions
About this article
Cite this article
Yu, J., Rao, P., Clark, S. et al. Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition. Nature 594, 448–453 (2021). https://doi.org/10.1038/s41586-021-03540-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03540-0
This article is cited by
-
GSG1L-containing AMPA receptor complexes are defined by their spatiotemporal expression, native interactome and allosteric sites
Nature Communications (2023)
-
Modulation of GluA2–γ5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2
Nature Structural & Molecular Biology (2023)
-
Automatic and accurate ligand structure determination guided by cryo-electron microscopy maps
Nature Communications (2023)
-
Enhanced TARP-γ8-PSD-95 coupling in excitatory neurons contributes to the rapid antidepressant-like action of ketamine in male mice
Nature Communications (2023)
-
Modulatory mechanisms of TARP γ8-selective AMPA receptor therapeutics
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.